Interferon regulatory factors (IRFs) repress transcription of the chicken ovalbumin gene
- PMID: 19341784
- PMCID: PMC2749989
- DOI: 10.1016/j.gene.2009.03.016
Interferon regulatory factors (IRFs) repress transcription of the chicken ovalbumin gene
Abstract
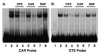
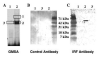
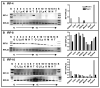
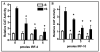
Although the ovalbumin (Ov) gene has served as a model to study tissue-specific, steroid hormone-induced gene expression in vertebrates for decades, the mechanisms responsible for regulating this gene remain elusive. Ov is repressed in non-oviduct tissue and in estrogen-deprived oviduct by a strong repressor site located from -130 to -100 and designated CAR for COUP-TF adjacent repressor. The goal of this study was to identify the CAR binding protein(s). A transcription factor database search revealed that a putative interferon-stimulated response element (ISRE), which binds interferon regulatory factors (IRFs), is located in this region. Gel mobility shift assays demonstrated that the protein(s) binding to the CAR site is recognized by an IRF antibody and that mutations in the ISRE abolish that binding. In hopes of identifying the IRF(s) responsible for the tissue-specific regulation of Ov, mRNA levels for IRFs-4, -8, and -10 were measured in seven tissues from chicks treated with or without estrogen. PCR experiments showed that both IRF-8 and -10 are expressed in all chick tissues tested whereas IRF-4 has a much more limited expression pattern. Transfection experiments with OvCAT (chloramphenicol acetyltransferase) reporter constructs demonstrated that both IRF-4 and IRF-10 are capable of repressing the Ov gene even in the presence of steroid hormones and that nucleotides in the ISRE are required for repression. These experiments indicate that the repressor activity associated with the CAR site is mediated by IRF family members and suggest that IRF members also repress Ov in non-oviduct tissues.
Figures



Similar articles
-
The COUP-adjacent repressor (CAR) element participates in the tissue-specific expression of the ovalbumin gene.Biochim Biophys Acta. 2000 Dec 15;1517(1):27-32. doi: 10.1016/s0167-4781(00)00241-4. Biochim Biophys Acta. 2000. PMID: 11118613
-
COUP-TF plays a dual role in the regulation of the ovalbumin gene.Biochemistry. 2000 Jul 25;39(29):8537-45. doi: 10.1021/bi0005862. Biochemistry. 2000. PMID: 10913260
-
Multiple promoter elements including a novel repressor site modulate expression of the chick ovalbumin gene.DNA Cell Biol. 1999 Feb;18(2):147-56. doi: 10.1089/104454999315538. DNA Cell Biol. 1999. PMID: 10073574
-
Direct Inhibition of IRF-Dependent Transcriptional Regulatory Mechanisms Associated With Disease.Front Immunol. 2019 May 24;10:1176. doi: 10.3389/fimmu.2019.01176. eCollection 2019. Front Immunol. 2019. PMID: 31178872 Free PMC article. Review.
-
IRF family of transcription factors as regulators of host defense.Annu Rev Immunol. 2001;19:623-55. doi: 10.1146/annurev.immunol.19.1.623. Annu Rev Immunol. 2001. PMID: 11244049 Review.
Cited by
-
Gene cloning and induced expression pattern of IRF4 and IRF10 in the Asian swamp eel (Monopterus albus).Dongwuxue Yanjiu. 2014 Sep;35(5):380-8. doi: 10.13918/j.issn.2095-8137.2014.5.380. Dongwuxue Yanjiu. 2014. PMID: 25297077 Free PMC article.
-
Study of the regulatory elements of the Ovalbumin gene promoter using CRISPR technology in chicken cells.J Biol Eng. 2023 Jul 17;17(1):46. doi: 10.1186/s13036-023-00367-3. J Biol Eng. 2023. PMID: 37461059 Free PMC article.
-
Molecular identification and functional characterization of IRF4 from common carp (Cyprinus carpio. L) in immune response: a negative regulator in the IFN and NF-κB signalling pathways.BMC Vet Res. 2022 Mar 17;18(1):106. doi: 10.1186/s12917-022-03205-8. BMC Vet Res. 2022. PMID: 35300694 Free PMC article.
-
Global characterization of interferon regulatory factor (IRF) genes in vertebrates: glimpse of the diversification in evolution.BMC Immunol. 2010 May 5;11:22. doi: 10.1186/1471-2172-11-22. BMC Immunol. 2010. PMID: 20444275 Free PMC article.
References
-
- Dadoune JP, Pawlak A, Alfonsi MF, Siffroi JP. Identification of transcripts by macroarrays, RT-PCR and in situ hybridization in human ejaculate spermatozoa. Mol. Human Reprod. 2005;11:133–140. - PubMed
-
- Dean DM, Jones PM, Sanders MM. Alterations in chromatin structure are implicated in the activation of the steroid hormone response unit in the ovalbumin gene. DNA Cell Biol. 2001;20:27–39. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

