Gene structures and processing of Arabidopsis thaliana HYL1-dependent pri-miRNAs
- PMID: 19304749
- PMCID: PMC2685107
- DOI: 10.1093/nar/gkp189
Gene structures and processing of Arabidopsis thaliana HYL1-dependent pri-miRNAs
Abstract
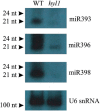
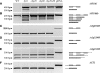
Arabidopsis thaliana HYL1 is a nuclear double-stranded RNA-binding protein involved in the maturation of pri-miRNAs. A quantitative real-time PCR platform for parallel quantification of 176 pri-miRNAs was used to reveal strong accumulation of 57 miRNA precursors in the hyl1 mutant that completely lacks HYL1 protein. This approach enabled us for the first time to pinpoint particular members of MIRNA family genes that require HYL1 activity for efficient maturation of their precursors. Moreover, the accumulation of miRNA precursors in the hyl1 mutant gave us the opportunity to carry out 3' and 5' RACE experiments which revealed that some of these precursors are of unexpected length. The alignment of HYL1-dependent miRNA precursors to A. thaliana genomic sequences indicated the presence of introns in 12 out of 20 genes studied. Some of the characterized intron-containing pri-miRNAs undergo alternative splicing such as exon skipping or usage of alternative 5' splice sites suggesting that this process plays a role in the regulation of miRNA biogenesis. In the hyl1 mutant intron-containing pri-miRNAs accumulate alongside spliced pri-miRNAs suggesting the recruitment of HYL1 into the miRNA precursor maturation pathway before their splicing occurs.
Figures



Similar articles
-
The N-terminal double-stranded RNA binding domains of Arabidopsis HYPONASTIC LEAVES1 are sufficient for pre-microRNA processing.Plant Cell. 2007 Mar;19(3):914-25. doi: 10.1105/tpc.106.048637. Epub 2007 Mar 2. Plant Cell. 2007. PMID: 17337628 Free PMC article.
-
The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis.RNA. 2006 Feb;12(2):206-12. doi: 10.1261/rna.2146906. RNA. 2006. PMID: 16428603 Free PMC article.
-
Intron Lariat RNA Inhibits MicroRNA Biogenesis by Sequestering the Dicing Complex in Arabidopsis.PLoS Genet. 2016 Nov 21;12(11):e1006422. doi: 10.1371/journal.pgen.1006422. eCollection 2016 Nov. PLoS Genet. 2016. PMID: 27870853 Free PMC article.
-
HYL1's multiverse: A journey through miRNA biogenesis and beyond canonical and non-canonical functions of HYL1.Curr Opin Plant Biol. 2024 Aug;80:102546. doi: 10.1016/j.pbi.2024.102546. Epub 2024 May 7. Curr Opin Plant Biol. 2024. PMID: 38718678 Review.
-
Orientation of Human Microprocessor on Primary MicroRNAs.Biochemistry. 2019 Jan 29;58(4):189-198. doi: 10.1021/acs.biochem.8b00944. Epub 2018 Dec 10. Biochemistry. 2019. PMID: 30481000 Review.
Cited by
-
MIR846 and MIR842 comprise a cistronic MIRNA pair that is regulated by abscisic acid by alternative splicing in roots of Arabidopsis.Plant Mol Biol. 2013 Mar;81(4-5):447-60. doi: 10.1007/s11103-013-0015-6. Epub 2013 Jan 23. Plant Mol Biol. 2013. PMID: 23341152 Free PMC article.
-
Arabidopsis microRNA expression regulation in a wide range of abiotic stress responses.Front Plant Sci. 2015 Jun 4;6:410. doi: 10.3389/fpls.2015.00410. eCollection 2015. Front Plant Sci. 2015. PMID: 26089831 Free PMC article.
-
The crosstalk between plant microRNA biogenesis factors and the spliceosome.Plant Signal Behav. 2013 Nov;8(11):e26955. doi: 10.4161/psb.26955. Epub 2013 Dec 3. Plant Signal Behav. 2013. PMID: 24300047 Free PMC article. Review.
-
Genomewide identification of genes involved in the potato response to drought indicates functional evolutionary conservation with Arabidopsis plants.Plant Biotechnol J. 2018 Feb;16(2):603-614. doi: 10.1111/pbi.12800. Epub 2017 Aug 14. Plant Biotechnol J. 2018. PMID: 28718511 Free PMC article.
-
Developmentally regulated expression and complex processing of barley pri-microRNAs.BMC Genomics. 2013 Jan 16;14:34. doi: 10.1186/1471-2164-14-34. BMC Genomics. 2013. PMID: 23324356 Free PMC article.
References
-
- Llave C, Xie Z, Kassachau KD, Carrington JC. Cleavage of Scarecrowlike mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. - PubMed
-
- Brodersen P, Sakvarelidze-Achard L, Brunn-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Vionnet O. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–1190. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

