Control of Pseudomonas aeruginosa AlgW protease cleavage of MucA by peptide signals and MucB
- PMID: 19298369
- PMCID: PMC2754073
- DOI: 10.1111/j.1365-2958.2009.06654.x
Control of Pseudomonas aeruginosa AlgW protease cleavage of MucA by peptide signals and MucB
Abstract
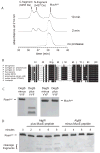
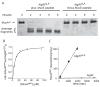
The ability of a pathogen to survive the defensive attacks of its host requires the detection of and response to perturbations in its own physiology. Activation of the extracytoplasmic stress response in the pathogen Pseudomonas aeruginosa results in higher tolerance against immune defences as well as in the production of alginate, a surface polysaccharide that also confers resistance to many host defences and antibiotic treatments. The alginate response is regulated by proteolytic cleavage of MucA, a transmembrane protein that inhibits the transcription factor AlgU, and by the periplasmic protein MucB. Here we show that specific peptides bind to the periplasmic AlgW protease and activate its cleavage of MucA. We demonstrate that tight binding of MucB to MucA strongly inhibits this cleavage. We also probe the roles of structural features of AlgW, including a key regulatory loop and its PDZ domain, in regulating substrate binding and cleavage.
Figures

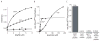


Similar articles
-
Overexpression of CupB5 activates alginate overproduction in Pseudomonas aeruginosa by a novel AlgW-dependent mechanism.Mol Microbiol. 2014 Aug;93(3):415-25. doi: 10.1111/mmi.12665. Epub 2014 Jul 6. Mol Microbiol. 2014. PMID: 24913916 Free PMC article.
-
In vivo and in vitro analyses of the role of the Prc protease in inducing mucoidy in Pseudomonas aeruginosa.J Bacteriol. 2024 Oct 24;206(10):e0022224. doi: 10.1128/jb.00222-24. Epub 2024 Sep 17. J Bacteriol. 2024. PMID: 39287400
-
Structural basis for the recognition of MucA by MucB and AlgU in Pseudomonas aeruginosa.FEBS J. 2019 Dec;286(24):4982-4994. doi: 10.1111/febs.14995. Epub 2019 Jul 19. FEBS J. 2019. PMID: 31297938
-
The algD promoter: regulation of alginate production by Pseudomonas aeruginosa in cystic fibrosis.Cell Mol Biol Res. 1993;39(4):371-6. Cell Mol Biol Res. 1993. PMID: 8312973 Review.
-
Iron uptake regulation in Pseudomonas aeruginosa.Biometals. 2009 Feb;22(1):15-22. doi: 10.1007/s10534-008-9193-0. Epub 2009 Jan 8. Biometals. 2009. PMID: 19130263 Review.
Cited by
-
Bacterial exopolysaccharides: biosynthesis pathways and engineering strategies.Front Microbiol. 2015 May 26;6:496. doi: 10.3389/fmicb.2015.00496. eCollection 2015. Front Microbiol. 2015. PMID: 26074894 Free PMC article. Review.
-
Bacterial alginate regulators and phage homologs repress CRISPR-Cas immunity.Nat Microbiol. 2020 May;5(5):679-687. doi: 10.1038/s41564-020-0691-3. Epub 2020 Mar 23. Nat Microbiol. 2020. PMID: 32203410 Free PMC article.
-
Bacterial sigma factors as targets for engineered or synthetic transcriptional control.Front Bioeng Biotechnol. 2014 Sep 3;2:33. doi: 10.3389/fbioe.2014.00033. eCollection 2014. Front Bioeng Biotechnol. 2014. PMID: 25232540 Free PMC article. Review.
-
Themes and variations in gene regulation by extracytoplasmic function (ECF) sigma factors.Curr Opin Microbiol. 2017 Apr;36:128-137. doi: 10.1016/j.mib.2017.05.004. Epub 2017 May 30. Curr Opin Microbiol. 2017. PMID: 28575802 Free PMC article. Review.
-
Draft genome sequence for Pseudomonas aeruginosa strain PAO579, a mucoid derivative of PAO381.J Bacteriol. 2012 Dec;194(23):6617. doi: 10.1128/JB.01406-12. J Bacteriol. 2012. PMID: 23144378 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

