The structure of an interdomain complex that regulates talin activity
- PMID: 19297334
- PMCID: PMC2685691
- DOI: 10.1074/jbc.M900078200
The structure of an interdomain complex that regulates talin activity
Abstract
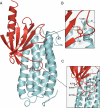
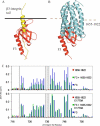
Talin is a large flexible rod-shaped protein that activates the integrin family of cell adhesion molecules and couples them to cytoskeletal actin. It exists in both globular and extended conformations, and an intramolecular interaction between the N-terminal F3 FERM subdomain and the C-terminal part of the talin rod contributes to an autoinhibited form of the molecule. Here, we report the solution structure of the primary F3 binding domain within the C-terminal region of the talin rod and use intermolecular nuclear Overhauser effects to determine the structure of the complex. The rod domain (residues 1655-1822) is an amphipathic five-helix bundle; Tyr-377 of F3 docks into a hydrophobic pocket at one end of the bundle, whereas a basic loop in F3 (residues 316-326) interacts with a cluster of acidic residues in the middle of helix 4. Mutation of Glu-1770 abolishes binding. The rod domain competes with beta3-integrin tails for binding to F3, and the structure of the complex suggests that the rod is also likely to sterically inhibit binding of the FERM domain to the membrane.
Figures

Similar articles
-
The talin rod IBS2 alpha-helix interacts with the beta3 integrin cytoplasmic tail membrane-proximal helix by establishing charge complementary salt bridges.J Biol Chem. 2008 Aug 29;283(35):24212-23. doi: 10.1074/jbc.M709704200. Epub 2008 Jun 23. J Biol Chem. 2008. PMID: 18577523 Free PMC article.
-
Integrin-bound talin head inhibits actin filament barbed-end elongation.J Biol Chem. 2018 Feb 16;293(7):2586-2596. doi: 10.1074/jbc.M117.808204. Epub 2017 Dec 24. J Biol Chem. 2018. PMID: 29276177 Free PMC article.
-
Structural basis for the interaction between the cytoplasmic domain of the hyaluronate receptor layilin and the talin F3 subdomain.J Mol Biol. 2008 Sep 26;382(1):112-26. doi: 10.1016/j.jmb.2008.06.087. Epub 2008 Jul 7. J Mol Biol. 2008. PMID: 18638481
-
Integrin connections to the cytoskeleton through talin and vinculin.Biochem Soc Trans. 2008 Apr;36(Pt 2):235-9. doi: 10.1042/BST0360235. Biochem Soc Trans. 2008. PMID: 18363566 Review.
-
Talin as a mechanosensitive signaling hub.J Cell Biol. 2018 Nov 5;217(11):3776-3784. doi: 10.1083/jcb.201808061. Epub 2018 Sep 25. J Cell Biol. 2018. PMID: 30254032 Free PMC article. Review.
Cited by
-
Promotive effect of Talin-1 protein on gastric cancer progression through PTK2-PXN-VCL-E-Cadherin-CAPN2-MAPK1 signaling axis.J Clin Lab Anal. 2020 Dec;34(12):e23555. doi: 10.1002/jcla.23555. Epub 2020 Sep 20. J Clin Lab Anal. 2020. PMID: 32951272 Free PMC article.
-
New isoform-specific monoclonal antibodies reveal different sub-cellular localisations for talin1 and talin2.Eur J Cell Biol. 2012 Mar;91(3):180-91. doi: 10.1016/j.ejcb.2011.12.003. Epub 2012 Feb 3. Eur J Cell Biol. 2012. PMID: 22306379 Free PMC article.
-
Kindlins, integrin activation and the regulation of talin recruitment to αIIbβ3.PLoS One. 2012;7(3):e34056. doi: 10.1371/journal.pone.0034056. Epub 2012 Mar 23. PLoS One. 2012. PMID: 22457811 Free PMC article.
-
The Structure of the talin head reveals a novel extended conformation of the FERM domain.Structure. 2010 Oct 13;18(10):1289-99. doi: 10.1016/j.str.2010.07.011. Structure. 2010. PMID: 20947018 Free PMC article.
-
Talin in mechanotransduction and mechanomemory at a glance.J Cell Sci. 2021 Oct 15;134(20):jcs258749. doi: 10.1242/jcs.258749. Epub 2021 Oct 28. J Cell Sci. 2021. PMID: 34708856 Free PMC article.
References
-
- Calderwood, D. A. (2004) J. Cell Sci. 117 657-666 - PubMed
-
- Critchley, D. R., and Gingras, A. R. (2008) J. Cell Sci. 121 1345-1347 - PubMed
-
- Tadokoro, S., Shattil, S. J., Eto, K., Tai, V., Liddington, R. C., de Pereda, J. M., Ginsberg, M. H., and Calderwood, D. A. (2003) Science 302 103-106 - PubMed
-
- Cram, E. J., Clark, S. G., and Schwarzbauer, J. E. (2003) J. Cell Sci. 116 3871-3878 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

