Human TRIM gene expression in response to interferons
- PMID: 19290053
- PMCID: PMC2654144
- DOI: 10.1371/journal.pone.0004894
Human TRIM gene expression in response to interferons
Abstract
Background: Tripartite motif (TRIM) proteins constitute a family of proteins that share a conserved tripartite architecture. The recent discovery of the anti-HIV activity of TRIM5alpha in primate cells has stimulated much interest in the potential role of TRIM proteins in antiviral activities and innate immunity.
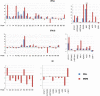
Principal findings: To test if TRIM genes are up-regulated during antiviral immune responses, we performed a systematic analysis of TRIM gene expression in human primary lymphocytes and monocyte-derived macrophages in response to interferons (IFNs, type I and II) or following FcgammaR-mediated activation of macrophages. We found that 27 of the 72 human TRIM genes are sensitive to IFN. Our analysis identifies 9 additional TRIM genes that are up-regulated by IFNs, among which only 3 have previously been found to display an antiviral activity. Also, we found 2 TRIM proteins, TRIM9 and 54, to be specifically up-regulated in FcgammaR-activated macrophages.
Conclusions: Our results present the first comprehensive TRIM gene expression analysis in primary human immune cells, and suggest the involvement of additional TRIM proteins in regulating host antiviral activities.
Conflict of interest statement
Figures





Similar articles
-
Interleukin 27, Similar to Interferons, Modulates Gene Expression of Tripartite Motif (TRIM) Family Members and Interferes with Mayaro Virus Replication in Human Macrophages.Viruses. 2024 Jun 20;16(6):996. doi: 10.3390/v16060996. Viruses. 2024. PMID: 38932287 Free PMC article.
-
Identification of a genomic reservoir for new TRIM genes in primate genomes.PLoS Genet. 2011 Dec;7(12):e1002388. doi: 10.1371/journal.pgen.1002388. Epub 2011 Dec 1. PLoS Genet. 2011. PMID: 22144910 Free PMC article.
-
TRIM protein-mediated regulation of inflammatory and innate immune signaling and its association with antiretroviral activity.J Virol. 2013 Jan;87(1):257-72. doi: 10.1128/JVI.01804-12. Epub 2012 Oct 17. J Virol. 2013. PMID: 23077300 Free PMC article.
-
TRIM family proteins and their emerging roles in innate immunity.Nat Rev Immunol. 2008 Nov;8(11):849-60. doi: 10.1038/nri2413. Nat Rev Immunol. 2008. PMID: 18836477 Free PMC article. Review.
-
Inhibition of retroviral replication by members of the TRIM protein family.Curr Top Microbiol Immunol. 2013;371:29-66. doi: 10.1007/978-3-642-37765-5_2. Curr Top Microbiol Immunol. 2013. PMID: 23686231 Review.
Cited by
-
Mammalian and Avian Host Cell Influenza A Restriction Factors.Viruses. 2021 Mar 22;13(3):522. doi: 10.3390/v13030522. Viruses. 2021. PMID: 33810083 Free PMC article. Review.
-
Exploring TRIM proteins' role in antiviral defense against influenza A virus and respiratory coronaviruses.Front Cell Infect Microbiol. 2024 Jul 15;14:1420854. doi: 10.3389/fcimb.2024.1420854. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39077432 Free PMC article. Review.
-
Antiviral TRIMs: friend or foe in autoimmune and autoinflammatory disease?Nat Rev Immunol. 2011 Aug 25;11(9):617-25. doi: 10.1038/nri3043. Nat Rev Immunol. 2011. PMID: 21866173
-
Genome editing of FTR42 improves zebrafish survival against virus infection by enhancing IFN immunity.iScience. 2024 Mar 12;27(4):109497. doi: 10.1016/j.isci.2024.109497. eCollection 2024 Apr 19. iScience. 2024. PMID: 38550983 Free PMC article.
-
TRIM5α Degradation via Autophagy Is Not Required for Retroviral Restriction.J Virol. 2016 Jan 13;90(7):3400-10. doi: 10.1128/JVI.03033-15. J Virol. 2016. PMID: 26764007 Free PMC article.
References
-
- Nisole S, Stoye JP, Saib A. TRIM family proteins: retroviral restriction and antiviral defence. Nat Rev Microbiol. 2005;3:799–808. - PubMed
-
- Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, et al. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

