Protection from isopeptidase-mediated deconjugation regulates paralog-selective sumoylation of RanGAP1
- PMID: 19285941
- PMCID: PMC2668917
- DOI: 10.1016/j.molcel.2009.02.008
Protection from isopeptidase-mediated deconjugation regulates paralog-selective sumoylation of RanGAP1
Abstract
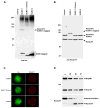
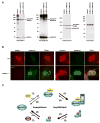
Vertebrates express three small ubiquitin-related modifiers (SUMO-1, SUMO-2, and SUMO-3) that are conjugated in part to unique subsets of proteins and, thereby, regulate distinct cellular processes. Mechanisms regulating paralog-selective sumoylation, however, remain poorly understood. Despite being equally well modified by SUMO-1 and SUMO-2 in vitro, RanGAP1 is selectively modified by SUMO-1 in vivo. We have found that this paralog-selective modification is determined at the level of deconjugation by isopeptidases. Our findings indicate that, relative to SUMO-2-modified RanGAP1, SUMO-1-modified RanGAP1 forms a more stable, higher affinity complex with the nucleoporin Nup358/RanBP2 that preferentially protects it from isopeptidases. By swapping residues in SUMO-1 and SUMO-2 responsible for Nup358/RanBP2 binding, or by manipulating isopeptidase expression levels, paralog-selective modification of RanGAP1 could be affected both in vitro and in vivo. Thus, protection from isopeptidases, through interactions with SUMO-binding proteins, represents an important mechanism defining paralog-selective sumoylation.
Figures




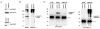
Similar articles
-
Alternative allosteric mechanisms can regulate the substrate and E2 in SUMO conjugation.J Mol Biol. 2011 Mar 4;406(4):620-30. doi: 10.1016/j.jmb.2010.12.044. Epub 2011 Jan 7. J Mol Biol. 2011. PMID: 21216249 Free PMC article.
-
Reconstitution of the Recombinant RanBP2 SUMO E3 Ligase Complex.Methods Mol Biol. 2016;1475:41-54. doi: 10.1007/978-1-4939-6358-4_3. Methods Mol Biol. 2016. PMID: 27631796
-
The RanBP2/RanGAP1*SUMO1/Ubc9 complex: a multisubunit E3 ligase at the intersection of sumoylation and the RanGTPase cycle.Nucleus. 2012 Sep-Oct;3(5):429-32. doi: 10.4161/nucl.21980. Epub 2012 Aug 27. Nucleus. 2012. PMID: 22925898 Free PMC article.
-
Performing in vitro sumoylation reactions using recombinant enzymes.Methods Mol Biol. 2009;497:187-99. doi: 10.1007/978-1-59745-566-4_12. Methods Mol Biol. 2009. PMID: 19107418 Review.
-
Ubiquitin-related modifier SUMO1 and nucleocytoplasmic transport.Traffic. 2002 Jun;3(6):381-7. doi: 10.1034/j.1600-0854.2002.30601.x. Traffic. 2002. PMID: 12010456 Review.
Cited by
-
Non-covalent Interaction With SUMO Enhances the Activity of Human Cytomegalovirus Protein IE1.Front Cell Dev Biol. 2021 May 13;9:662522. doi: 10.3389/fcell.2021.662522. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34055792 Free PMC article.
-
SUMO binding by the Epstein-Barr virus protein kinase BGLF4 is crucial for BGLF4 function.J Virol. 2012 May;86(10):5412-21. doi: 10.1128/JVI.00314-12. Epub 2012 Mar 7. J Virol. 2012. PMID: 22398289 Free PMC article.
-
Two distinct sites in Nup153 mediate interaction with the SUMO proteases SENP1 and SENP2.Nucleus. 2012 Jul 1;3(4):349-58. doi: 10.4161/nucl.20822. Epub 2012 Jun 12. Nucleus. 2012. PMID: 22688647 Free PMC article.
-
Alternative allosteric mechanisms can regulate the substrate and E2 in SUMO conjugation.J Mol Biol. 2011 Mar 4;406(4):620-30. doi: 10.1016/j.jmb.2010.12.044. Epub 2011 Jan 7. J Mol Biol. 2011. PMID: 21216249 Free PMC article.
-
The dynamics and mechanism of SUMO chain deconjugation by SUMO-specific proteases.J Biol Chem. 2011 Mar 25;286(12):10238-47. doi: 10.1074/jbc.M110.205153. Epub 2011 Jan 19. J Biol Chem. 2011. PMID: 21247896 Free PMC article.
References
-
- Alkuraya FS, Saadi I, Lund JJ, Turbe-Doan A, Morton CC, Maas RL. SUMO1 haploinsufficiency leads to cleft lip and palate. Science. 2006;313:1751. - PubMed
-
- Bailey D, O’Hare P. Characterization of the localization and proteolytic activity of the SUMO-specific protease, SENP1. J Biol Chem. 2004;279:692–703. - PubMed
-
- Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell. 2002;108:345–356. - PubMed
-
- Evdokimov E, Sharma P, Lockett SJ, Lualdi M, Kuehn MR. Loss of SUMO1 in mice affects RanGAP1 localization and formation of PML nuclear bodies, but is not lethal as it can be compensated by SUMO2 or SUMO3. J Cell Sci. 2008;121:4106–4113. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

