Characterization of a myristoylated, monomeric HIV Gag protein
- PMID: 19285328
- PMCID: PMC2683466
- DOI: 10.1016/j.virol.2009.02.037
Characterization of a myristoylated, monomeric HIV Gag protein
Abstract
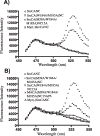
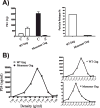
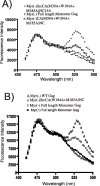
The process of HIV assembly requires extensive homomultimerization of the Gag polyprotein on cellular membranes to generate the nascent particle bud. Here we generated a full-length, monomeric Gag polyprotein bearing mutations that eliminated multimerization in living cells as indicated by fluorescence resonance energy transfer (FRET). Monomeric Gag resembled non-myristoylated Gag in its weak membrane binding characteristics and lack of association with detergent-resistant membranes (DRMs or lipid rafts). Monomeric Gag failed to assemble virus-like particles, but was inefficiently rescued into particles by wildtype Gag through the influence of the matrix domain. The subcellular distribution of monomeric Gag was remarkably different than either non-myristoylated Gag or wildtype Gag. Monomeric Gag was found on intracellular membranes and at the plasma membrane, where it highlighted plasma membrane extensions and ruffles. This study indicates that monomeric Gag can traffic to assembly sites in the cell, where it interacts weakly with membranes.
Figures


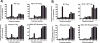


Similar articles
-
Quantitative fluorescence resonance energy transfer microscopy analysis of the human immunodeficiency virus type 1 Gag-Gag interaction: relative contributions of the CA and NC domains and membrane binding.J Virol. 2009 Jul;83(14):7322-36. doi: 10.1128/JVI.02545-08. Epub 2009 Apr 29. J Virol. 2009. PMID: 19403686 Free PMC article.
-
Myristoylation is required for human immunodeficiency virus type 1 Gag-Gag multimerization in mammalian cells.J Virol. 2007 Dec;81(23):12899-910. doi: 10.1128/JVI.01280-07. Epub 2007 Sep 19. J Virol. 2007. PMID: 17881447 Free PMC article.
-
Role of the nucleocapsid domain in HIV-1 Gag oligomerization and trafficking to the plasma membrane: a fluorescence lifetime imaging microscopy investigation.J Mol Biol. 2015 Mar 27;427(6 Pt B):1480-1494. doi: 10.1016/j.jmb.2015.01.015. Epub 2015 Jan 30. J Mol Biol. 2015. PMID: 25644662
-
How HIV-1 Gag Manipulates Its Host Cell Proteins: A Focus on Interactors of the Nucleocapsid Domain.Viruses. 2020 Aug 13;12(8):888. doi: 10.3390/v12080888. Viruses. 2020. PMID: 32823718 Free PMC article. Review.
-
New insights into HIV assembly and trafficking.Physiology (Bethesda). 2011 Aug;26(4):236-51. doi: 10.1152/physiol.00051.2010. Physiology (Bethesda). 2011. PMID: 21841072 Free PMC article. Review.
Cited by
-
Post-translational intracellular trafficking determines the type of immune response elicited by DNA vaccines expressing Gag antigen of Human Immunodeficiency Virus Type 1 (HIV-1).Hum Vaccin Immunother. 2013 Oct;9(10):2095-102. doi: 10.4161/hv.26009. Epub 2013 Aug 13. Hum Vaccin Immunother. 2013. PMID: 23941868 Free PMC article.
-
Relationships between plasma membrane microdomains and HIV-1 assembly.Biol Cell. 2010 Mar 25;102(6):335-50. doi: 10.1042/BC20090165. Biol Cell. 2010. PMID: 20356318 Free PMC article. Review.
-
FLIM FRET technology for drug discovery: automated multiwell-plate high-content analysis, multiplexed readouts and application in situ.Chemphyschem. 2011 Feb 25;12(3):609-26. doi: 10.1002/cphc.201000874. Epub 2011 Feb 17. Chemphyschem. 2011. PMID: 21337485 Free PMC article.
-
HIV-1 Gag as an Antiviral Target: Development of Assembly and Maturation Inhibitors.Curr Top Med Chem. 2016;16(10):1154-66. doi: 10.2174/1568026615666150902102143. Curr Top Med Chem. 2016. PMID: 26329615 Free PMC article. Review.
-
Clustering and mobility of HIV-1 Env at viral assembly sites predict its propensity to induce cell-cell fusion.J Virol. 2013 Jul;87(13):7516-25. doi: 10.1128/JVI.00790-13. Epub 2013 May 1. J Virol. 2013. PMID: 23637402 Free PMC article.
References
-
- Briggs JA, Johnson MC, Simon MN, Fuller SD, Vogt VM. Cryo-electron microscopy reveals conserved and divergent features of gag packing in immature particles of Rous sarcoma virus and human immunodeficiency virus. J Mol Biol. 2006;355 (1):157–68. - PubMed
-
- Briggs JA, Simon MN, Gross I, Krausslich HG, Fuller SD, Vogt VM, Johnson MC. The stoichiometry of Gag protein in HIV-1. Nat Struct Mol Biol. 2004;11 (7):672–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

