Evolutionary and biophysical relationships among the papillomavirus E2 proteins
- PMID: 19273107
- PMCID: PMC3705561
- DOI: 10.2741/3285
Evolutionary and biophysical relationships among the papillomavirus E2 proteins
Abstract
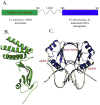
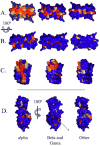
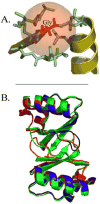
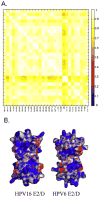
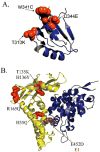
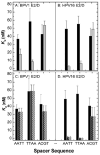
Infection by human papillomavirus (HPV) may result in clinical conditions ranging from benign warts to invasive cancer. The HPV E2 protein represses oncoprotein transcription and is required for viral replication. HPV E2 binds to palindromic DNA sequences of highly conserved four base pair sequences flanking an identical length variable 'spacer'. E2 proteins directly contact the conserved but not the spacer DNA. Variation in naturally occurring spacer sequences results in differential protein affinity that is dependent on their sensitivity to the spacer DNA's unique conformational and/or dynamic properties. This article explores the biophysical character of this core viral protein with the goal of identifying characteristics that associated with risk of virally caused malignancy. The amino acid sequence, 3d structure and electrostatic features of the E2 protein DNA binding domain are highly conserved; specific interactions with DNA binding sites have also been conserved. In contrast, the E2 protein's transactivation domain does not have extensive surfaces of highly conserved residues. Rather, regions of high conservation are localized to small surface patches. Implications to cancer biology are discussed.
Figures



Similar articles
-
DNA structure and flexibility in the sequence-specific binding of papillomavirus E2 proteins.J Mol Biol. 1998 Mar 6;276(4):809-18. doi: 10.1006/jmbi.1997.1578. J Mol Biol. 1998. PMID: 9500925
-
Indirect readout of DNA sequence by papillomavirus E2 proteins depends upon net cation uptake.J Mol Biol. 2006 Apr 21;358(1):224-40. doi: 10.1016/j.jmb.2006.01.093. Epub 2006 Feb 14. J Mol Biol. 2006. PMID: 16513133
-
Comparison of the structure and DNA-binding properties of the E2 proteins from an oncogenic and a non-oncogenic human papillomavirus.J Mol Biol. 2003 Dec 12;334(5):979-91. doi: 10.1016/j.jmb.2003.10.009. J Mol Biol. 2003. PMID: 14643661
-
Identification of single amino acids in the human papillomavirus 11 E2 protein critical for the transactivation or replication functions.Virology. 1998 Feb 15;241(2):312-22. doi: 10.1006/viro.1997.8941. Virology. 1998. PMID: 9499806
-
The E8 repression domain can replace the E2 transactivation domain for growth inhibition of HeLa cells by papillomavirus E2 proteins.Int J Cancer. 2007 Nov 15;121(10):2284-92. doi: 10.1002/ijc.22907. Int J Cancer. 2007. PMID: 17583574
Cited by
-
Human papillomavirus molecular biology and disease association.Rev Med Virol. 2015 Mar;25 Suppl 1(Suppl Suppl 1):2-23. doi: 10.1002/rmv.1822. Rev Med Virol. 2015. PMID: 25752814 Free PMC article. Review.
-
Detection and analysis of human papillomavirus (HPV) DNA in breast cancer patients by an effective method of HPV capture.PLoS One. 2014 Mar 10;9(3):e90343. doi: 10.1371/journal.pone.0090343. eCollection 2014. PLoS One. 2014. PMID: 24614680 Free PMC article.
-
In vivo Antitumor Effect of an HPV-specific Promoter driving IL-12 Expression in an HPV 16-positive Murine Model of Cervical Cancer.J Cancer. 2016 Sep 27;7(14):1950-1959. doi: 10.7150/jca.15536. eCollection 2016. J Cancer. 2016. PMID: 27877210 Free PMC article.
-
Whole genome sequencing and evolutionary analysis of human papillomavirus type 16 in central China.PLoS One. 2012;7(5):e36577. doi: 10.1371/journal.pone.0036577. Epub 2012 May 4. PLoS One. 2012. PMID: 22574185 Free PMC article.
-
E2 proteins of high risk human papillomaviruses down-modulate STING and IFN-κ transcription in keratinocytes.PLoS One. 2014 Mar 10;9(3):e91473. doi: 10.1371/journal.pone.0091473. eCollection 2014. PLoS One. 2014. PMID: 24614210 Free PMC article.
References
-
- de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. Classification of papillomaviruses. Virology. 2004;324(1):17–27. - PubMed
-
- Kadish AS, Burk RD, Kress Y, Calderin S, Romney SL. Human papillomaviruses of different types in precancerous lesions of the uterine cervix: Histologic, immunocytochemical and ultrastructural studies. Hum Pathol. 1986;17(4):384–92. - PubMed
-
- Van Ranst M, Fuse A, Fiten P, Beuken E, Pfister H, Burk RD, Opdenakker G. Human papillomavirus type 13 and pygmy chimpanzee papillomavirus type 1: Comparison of the genome organizations. Virology. 1992;190:587–596. - PubMed
-
- Munoz N, Bosch FX, de Sanjose S, Herrero R, Castellsague X, Shah KV, Snijders PJ, Meijer CJ. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348(6):518–27. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

