MERIT40 facilitates BRCA1 localization and DNA damage repair
- PMID: 19261748
- PMCID: PMC2661609
- DOI: 10.1101/gad.1770609
MERIT40 facilitates BRCA1 localization and DNA damage repair
Abstract
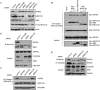
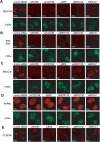
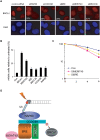
The product of breast cancer susceptibility gene 1, BRCA1, plays pivotal roles in the maintenance of genomic integrity. Mounting evidence indicates that BRCA1 associates with many proteins or protein complexes to regulate diverse processes important for the cellular response to DNA damage. One of these complexes, which mediates the accumulation of BRCA1 at sites of DNA breaks, involves the ubiquitin-binding motif (UIM)-containing protein RAP80, a coiled-coil domain protein CCDC98/Abraxas, and a deubiquitinating enzyme BRCC36. Here we describe the characterization of a novel component of this complex, MERIT40 (Mediator of Rap80 Interactions and Targeting 40 kd), which together with an adaptor protein BRE/BRCC45, enforces the BRCA1-dependent DNA damage response. MERIT40 is assembled into this RAP80/CCDC98-containing complex via its direct interaction with BRE/BRCC45. Importantly, MERIT40 regulates BRCA1 retention at DNA breaks and checkpoint function primarily via a role in maintaining the stability of BRE and this five-subunit protein complex at sites of DNA damage. Together, our study reveals that a stable complex containing MERIT40 acts early in DNA damage response and regulates damage-dependent BRCA1 localization.
Figures


Similar articles
-
MERIT40 controls BRCA1-Rap80 complex integrity and recruitment to DNA double-strand breaks.Genes Dev. 2009 Mar 15;23(6):740-54. doi: 10.1101/gad.1739609. Epub 2009 Mar 4. Genes Dev. 2009. PMID: 19261746 Free PMC article.
-
NBA1/MERIT40 and BRE interaction is required for the integrity of two distinct deubiquitinating enzyme BRCC36-containing complexes.J Biol Chem. 2011 Apr 1;286(13):11734-45. doi: 10.1074/jbc.M110.200857. Epub 2011 Jan 31. J Biol Chem. 2011. PMID: 21282113 Free PMC article.
-
Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage.Proc Natl Acad Sci U S A. 2007 Dec 26;104(52):20759-63. doi: 10.1073/pnas.0710061104. Epub 2007 Dec 5. Proc Natl Acad Sci U S A. 2007. PMID: 18077395 Free PMC article.
-
RAP80 and RNF8, key players in the recruitment of repair proteins to DNA damage sites.Cancer Lett. 2008 Nov 28;271(2):179-90. doi: 10.1016/j.canlet.2008.04.046. Epub 2008 Jun 11. Cancer Lett. 2008. PMID: 18550271 Free PMC article. Review.
-
Factors forming the BRCA1-A complex orchestrate BRCA1 recruitment to the sites of DNA damage.Acta Biochim Biophys Sin (Shanghai). 2016 Jul;48(7):658-64. doi: 10.1093/abbs/gmw047. Epub 2016 Jun 20. Acta Biochim Biophys Sin (Shanghai). 2016. PMID: 27325824 Review.
Cited by
-
BRE Promotes Esophageal Squamous Cell Carcinoma Growth by Activating AKT Signaling.Front Oncol. 2020 Aug 11;10:1407. doi: 10.3389/fonc.2020.01407. eCollection 2020. Front Oncol. 2020. PMID: 32850455 Free PMC article.
-
Loss of BRCA1-A complex function in RAP80 null tumor cells.PLoS One. 2012;7(7):e40406. doi: 10.1371/journal.pone.0040406. Epub 2012 Jul 6. PLoS One. 2012. PMID: 22792303 Free PMC article.
-
RAP80 protein is important for genomic stability and is required for stabilizing BRCA1-A complex at DNA damage sites in vivo.J Biol Chem. 2012 Jun 29;287(27):22919-26. doi: 10.1074/jbc.M112.351007. Epub 2012 Apr 25. J Biol Chem. 2012. PMID: 22539352 Free PMC article.
-
Double strand break repair functions of histone H2AX.Mutat Res. 2013 Oct;750(1-2):5-14. doi: 10.1016/j.mrfmmm.2013.07.007. Epub 2013 Jul 31. Mutat Res. 2013. PMID: 23916969 Free PMC article. Review.
-
Expression of the BRCA1 complex member BRE predicts disease free survival in breast cancer.Breast Cancer Res Treat. 2012 Aug;135(1):125-33. doi: 10.1007/s10549-012-2122-5. Epub 2012 Jun 16. Breast Cancer Res Treat. 2012. PMID: 22706632 Free PMC article.
References
-
- Baer R., Ludwig T. The BRCA1/BARD1 heterodimer, a tumor suppressor complex with ubiquitin E3 ligase activity. Curr. Opin. Genet. Dev. 2002;12:86–91. - PubMed
-
- Balakirev M.Y., Wilkinson K.D. OTU takes the chains OUT. Nat. Chem. Biol. 2008;4:227–228. - PubMed
-
- Bennett E.J., Harper J.W. DNA damage: Ubiquitin marks the spot. Nat. Struct. Mol. Biol. 2008;15:20–22. - PubMed
-
- Boulton S.J. BRCA1-mediated ubiquitylation. Cell Cycle. 2006;5:1481–1486. - PubMed
-
- Cantor S.B., Bell D.W., Ganesan S., Kass E.M., Drapkin R., Grossman S., Wahrer D.C., Sgroi D.C., Lane W.S., Haber D.A., et al. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001;105:149–160. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
