Analysis of lentiviral vector integration in HIV+ study subjects receiving autologous infusions of gene modified CD4+ T cells
- PMID: 19259065
- PMCID: PMC2835137
- DOI: 10.1038/mt.2009.16
Analysis of lentiviral vector integration in HIV+ study subjects receiving autologous infusions of gene modified CD4+ T cells
Abstract
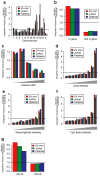
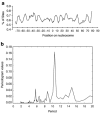
Lentiviral vector-based gene therapy has been used to target the human immunodeficiency virus (HIV) using an antisense env payload. We have analyzed lentiviral-vector integration sites from three treated individuals. We compared integration sites from the ex vivo vector-transduced CD4+ cell products to sites from cells recovered at several times after infusion. Integration sites were analyzed using 454 pyrosequencing, yielding a total of 7,782 unique integration sites from the ex vivo product and 237 unique sites from cells recovered after infusion. Integrated vector copies in both data sets were found to be strongly enriched within active genes and near epigenetic marks associated with active transcription units. Analysis of integration relative to nucleosome structure on target DNA indicated favoring of integration in outward facing DNA major grooves on the nucleosome surface. There was no indication that growth of transduced cells after infusion resulted in enrichment for integration sites near proto-oncogene 5'-ends or within tumor suppressor genes. Thus, this first look at the longitudinal evolution of cells transduced with a lentiviral vector after infusion of gene modified CD4+ cells provided no evidence for abnormal expansions of cells due to vector-mediated insertional activation of proto-oncogenes.
Figures



Comment in
-
Dotting the I's and crossing the T's: integration analyses in transduced patient T cells.Mol Ther. 2009 May;17(5):756-7. doi: 10.1038/mt.2009.75. Mol Ther. 2009. PMID: 19404325 Free PMC article. No abstract available.
Similar articles
-
Antiviral effects of autologous CD4 T cells genetically modified with a conditionally replicating lentiviral vector expressing long antisense to HIV.Blood. 2013 Feb 28;121(9):1524-33. doi: 10.1182/blood-2012-07-447250. Epub 2012 Dec 20. Blood. 2013. PMID: 23264589 Free PMC article. Clinical Trial.
-
Dotting the I's and crossing the T's: integration analyses in transduced patient T cells.Mol Ther. 2009 May;17(5):756-7. doi: 10.1038/mt.2009.75. Mol Ther. 2009. PMID: 19404325 Free PMC article. No abstract available.
-
Comparison of HIV-derived lentiviral and MLV-based gammaretroviral vector integration sites in primate repopulating cells.Mol Ther. 2007 Jul;15(7):1356-65. doi: 10.1038/sj.mt.6300159. Epub 2007 Apr 17. Mol Ther. 2007. PMID: 17440443
-
Lentiviral vectors for gene therapy of HIV-1 infection.Curr Gene Ther. 2002 Feb;2(1):23-43. doi: 10.2174/1566523023348165. Curr Gene Ther. 2002. PMID: 12108972 Review.
-
[Progress in improvement of lentiviral vectors' transcriptional read-through].Sheng Wu Gong Cheng Xue Bao. 2011 Nov;27(11):1541-8. Sheng Wu Gong Cheng Xue Bao. 2011. PMID: 22393708 Review. Chinese.
Cited by
-
Stem cell gene therapy for HIV: strategies to inhibit viral entry and replication.Curr HIV/AIDS Rep. 2015 Mar;12(1):79-87. doi: 10.1007/s11904-014-0242-8. Curr HIV/AIDS Rep. 2015. PMID: 25578054 Review.
-
Novel cell and gene therapies for HIV.Cold Spring Harb Perspect Med. 2012 Oct 1;2(10):a007179. doi: 10.1101/cshperspect.a007179. Cold Spring Harb Perspect Med. 2012. PMID: 23028130 Free PMC article. Review.
-
Safety and efficacy of axicabtagene ciloleucel in refractory large B-cell lymphomas.Ther Adv Hematol. 2020 Jan 29;11:2040620720902899. doi: 10.1177/2040620720902899. eCollection 2020. Ther Adv Hematol. 2020. PMID: 32064069 Free PMC article. Review.
-
Cellular unfolded protein response against viruses used in gene therapy.Front Microbiol. 2014 May 26;5:250. doi: 10.3389/fmicb.2014.00250. eCollection 2014. Front Microbiol. 2014. PMID: 24904562 Free PMC article. Review.
-
Antiviral effects of autologous CD4 T cells genetically modified with a conditionally replicating lentiviral vector expressing long antisense to HIV.Blood. 2013 Feb 28;121(9):1524-33. doi: 10.1182/blood-2012-07-447250. Epub 2012 Dec 20. Blood. 2013. PMID: 23264589 Free PMC article. Clinical Trial.
References
-
- Cavazzana-Calvo M, Hacein-Bey S, de Saint Basile G, Gross F, Yvon E, Nusbaum P, et al. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science. 2000;288:669–672. - PubMed
-
- Hacein-Bey-Abina S, Le Deist F, Carlier F, Bouneaud C, Hue C, De Villartay JP, et al. Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. N Engl J Med. 2002;346:1185–1193. - PubMed
-
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. - PubMed
-
- Hacein-Bey-Abina S, von Kalle C, Schmidt M, Le Deist F, Wulffraat N, McIntyre E, et al. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N Engl J Med. 2003;348:255–256. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

