Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance
- PMID: 19258535
- PMCID: PMC2650891
- DOI: 10.1128/MMBR.00034-08
Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance
Abstract
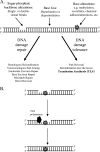
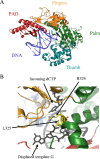
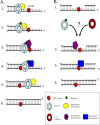
DNA repair and DNA damage tolerance machineries are crucial to overcome the vast array of DNA damage that a cell encounters during its lifetime. In this review, we summarize the current state of knowledge about the eukaryotic DNA damage tolerance pathway translesion synthesis (TLS), a process in which specialized DNA polymerases replicate across from DNA lesions. TLS aids in resistance to DNA damage, presumably by restarting stalled replication forks or filling in gaps that remain in the genome due to the presence of DNA lesions. One consequence of this process is the potential risk of introducing mutations. Given the role of these translesion polymerases in mutagenesis, we discuss the significant regulatory mechanisms that control the five known eukaryotic translesion polymerases: Rev1, Pol zeta, Pol kappa, Pol eta, and Pol iota.
Figures

Similar articles
-
Filling gaps in translesion DNA synthesis in human cells.Mutat Res Genet Toxicol Environ Mutagen. 2018 Dec;836(Pt B):127-142. doi: 10.1016/j.mrgentox.2018.02.004. Epub 2018 Feb 23. Mutat Res Genet Toxicol Environ Mutagen. 2018. PMID: 30442338 Review.
-
Structural basis of Rev1-mediated assembly of a quaternary vertebrate translesion polymerase complex consisting of Rev1, heterodimeric polymerase (Pol) ζ, and Pol κ.J Biol Chem. 2012 Sep 28;287(40):33836-46. doi: 10.1074/jbc.M112.394841. Epub 2012 Aug 2. J Biol Chem. 2012. PMID: 22859295 Free PMC article.
-
Translesion Synthesis of the N(2)-2'-Deoxyguanosine Adduct of the Dietary Mutagen IQ in Human Cells: Error-Free Replication by DNA Polymerase κ and Mutagenic Bypass by DNA Polymerases η, ζ, and Rev1.Chem Res Toxicol. 2016 Sep 19;29(9):1549-59. doi: 10.1021/acs.chemrestox.6b00221. Epub 2016 Aug 17. Chem Res Toxicol. 2016. PMID: 27490094 Free PMC article.
-
Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis.EMBO J. 2003 Dec 15;22(24):6621-30. doi: 10.1093/emboj/cdg626. EMBO J. 2003. PMID: 14657033 Free PMC article.
-
Eukaryotic TLS polymerases.Postepy Hig Med Dosw (Online). 2016 May 21;70(0):522-33. doi: 10.5604/17322693.1202481. Postepy Hig Med Dosw (Online). 2016. PMID: 27333922 Review.
Cited by
-
Regulation of translesion DNA synthesis: Posttranslational modification of lysine residues in key proteins.DNA Repair (Amst). 2015 May;29:166-79. doi: 10.1016/j.dnarep.2015.02.011. Epub 2015 Feb 18. DNA Repair (Amst). 2015. PMID: 25743599 Free PMC article. Review.
-
DNA polymerase zeta generates clustered mutations during bypass of endogenous DNA lesions in Saccharomyces cerevisiae.Environ Mol Mutagen. 2012 Dec;53(9):777-86. doi: 10.1002/em.21728. Epub 2012 Sep 11. Environ Mol Mutagen. 2012. PMID: 22965922 Free PMC article.
-
Targeting DNA Repair, Cell Cycle, and Tumor Microenvironment in B Cell Lymphoma.Cells. 2020 Oct 14;9(10):2287. doi: 10.3390/cells9102287. Cells. 2020. PMID: 33066395 Free PMC article. Review.
-
ZRANB3 is a structure-specific ATP-dependent endonuclease involved in replication stress response.Genes Dev. 2012 Jul 15;26(14):1558-72. doi: 10.1101/gad.193516.112. Epub 2012 Jul 3. Genes Dev. 2012. PMID: 22759634 Free PMC article.
-
Mechanism of RNA polymerase II bypass of oxidative cyclopurine DNA lesions.Proc Natl Acad Sci U S A. 2015 Feb 3;112(5):E410-9. doi: 10.1073/pnas.1415186112. Epub 2015 Jan 20. Proc Natl Acad Sci U S A. 2015. PMID: 25605892 Free PMC article.
References
-
- Albertella, M. R., C. M. Green, A. R. Lehmann, and M. J. O'Connor. 2005. A role for polymerase eta in the cellular tolerance to cisplatin-induced damage. Cancer Res. 659799-9806. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

