Tristetraprolin mediates interferon-gamma mRNA decay
- PMID: 19258311
- PMCID: PMC2670126
- DOI: 10.1074/jbc.M901229200
Tristetraprolin mediates interferon-gamma mRNA decay
Abstract
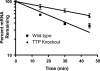
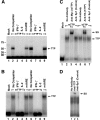
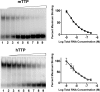
Tristetraprolin (TTP) regulates expression at the level of mRNA decay of several cytokines, including the T cell-specific cytokine, interleukin-2. We performed experiments to determine whether another T cell-specific cytokine, interferon-gamma (IFN-gamma), is also regulated by TTP and found that T cell receptor-activated T cells from TTP knock-out mice overproduced IFN-gamma mRNA and protein compared with activated T cells from wild-type mice. The half-life of IFN-gamma mRNA was 23 min in anti-CD3-stimulated T cells from wild-type mice, whereas it was 51 min in anti-CD3-stimulated T cells from TTP knock-out mice, suggesting that the overexpression of IFN-gamma mRNA in TTP knock-out mice was due to stabilization of IFN-gamma mRNA. Insertion of a 70-nucleotide AU-rich sequence from the murine IFN-gamma 3'-untranslated region, which contained a high affinity binding site for TTP, into the 3'-untranslated region of a beta-globin reporter transcript conferred TTP-dependent destabilization on the beta-globin transcript. Together these results suggest that TTP binds to a functional AU-rich element in the 3'-untranslated region of IFN-gamma mRNA and mediates rapid degradation of the IFN-gamma transcript. Thus, TTP plays an important role in turning off IFN-gamma expression at the appropriate time during an immune response.
Figures
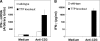

Similar articles
-
The roles of TTP and BRF proteins in regulated mRNA decay.Wiley Interdiscip Rev RNA. 2011 Jan-Feb;2(1):42-57. doi: 10.1002/wrna.28. Wiley Interdiscip Rev RNA. 2011. PMID: 21278925 Free PMC article. Review.
-
Tristetraprolin down-regulates IL-2 gene expression through AU-rich element-mediated mRNA decay.J Immunol. 2005 Jan 15;174(2):953-61. doi: 10.4049/jimmunol.174.2.953. J Immunol. 2005. PMID: 15634918
-
Tristetraprolin recruits functional mRNA decay complexes to ARE sequences.J Cell Biochem. 2007 Apr 15;100(6):1477-92. doi: 10.1002/jcb.21130. J Cell Biochem. 2007. PMID: 17133347
-
Posttranscriptional regulation of IL-23 expression by IFN-gamma through tristetraprolin.J Immunol. 2011 Jun 1;186(11):6454-64. doi: 10.4049/jimmunol.1002672. Epub 2011 Apr 22. J Immunol. 2011. PMID: 21515794 Free PMC article.
-
Control of mRNA decay by phosphorylation of tristetraprolin.Biochem Soc Trans. 2008 Jun;36(Pt 3):491-6. doi: 10.1042/BST0360491. Biochem Soc Trans. 2008. PMID: 18481987 Review.
Cited by
-
A single-cell transcriptional gradient in human cutaneous memory T cells restricts Th17/Tc17 identity.Cell Rep Med. 2022 Aug 16;3(8):100715. doi: 10.1016/j.xcrm.2022.100715. Cell Rep Med. 2022. PMID: 35977472 Free PMC article.
-
The roles of TTP and BRF proteins in regulated mRNA decay.Wiley Interdiscip Rev RNA. 2011 Jan-Feb;2(1):42-57. doi: 10.1002/wrna.28. Wiley Interdiscip Rev RNA. 2011. PMID: 21278925 Free PMC article. Review.
-
Effect of the p53-tristetraprolin-stathmin-1 pathway on trophoblasts at maternal-fetal interface.PLoS One. 2017 Jun 28;12(6):e0179852. doi: 10.1371/journal.pone.0179852. eCollection 2017. PLoS One. 2017. PMID: 28658321 Free PMC article.
-
RNA-Binding Protein ZFP36L2 Downregulates Helios Expression and Suppresses the Function of Regulatory T Cells.Front Immunol. 2020 Jun 23;11:1291. doi: 10.3389/fimmu.2020.01291. eCollection 2020. Front Immunol. 2020. PMID: 32655569 Free PMC article.
-
Dance with the Devil: Stress Granules and Signaling in Antiviral Responses.Viruses. 2020 Sep 4;12(9):984. doi: 10.3390/v12090984. Viruses. 2020. PMID: 32899736 Free PMC article. Review.
References
-
- Boehm, U., Klamp, T., Groot, M., and Howard, J. C. (1997) Annu. Rev. Immunol. 15, 749-795 - PubMed
-
- Schroder, K., Hertzog, P. J., Ravasi, T., and Hume, D. A. (2004) J. Leukoc. Biol. 75, 163-189 - PubMed
-
- Muller, U., Steinhoff, U., Reis, L. F., Hemmi, S., Pavlovic, J., Zinkernagel, R. M., and Aguet, M. (1994) Science 264, 1918-1921 - PubMed
-
- Huang, S., Hendriks, W., Althage, A., Hemmi, S., Bluethmann, H., Kamijo, R., Vilcek, J., Zinkernagel, R. M., and Aguet, M. (1993) Science 259, 1742-1745 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

