Transcriptional scaffolds for heterochromatin assembly
- PMID: 19239883
- PMCID: PMC6309854
- DOI: 10.1016/j.cell.2009.02.004
Transcriptional scaffolds for heterochromatin assembly
Abstract
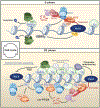
Heterochromatin is dynamically regulated during the cell cycle and in response to developmental signals. Recent findings from diverse systems suggest an extensive role for transcription in the assembly of heterochromatin, highlighting the emerging theme that transcription and noncoding RNAs can provide the initial scaffold for the formation of heterochromatin, which serves as a versatile recruiting platform for diverse factors involved in many cellular processes.
Figures
Similar articles
-
Noncoding RNAs and the borders of heterochromatin.Wiley Interdiscip Rev RNA. 2014 Nov-Dec;5(6):835-47. doi: 10.1002/wrna.1249. Epub 2014 Jul 9. Wiley Interdiscip Rev RNA. 2014. PMID: 25044367 Free PMC article. Review.
-
[Heterochromatin as a dynamic higher order chromatin structure].Tanpakushitsu Kakusan Koso. 2009 Mar;54(4 Suppl):488-95. Tanpakushitsu Kakusan Koso. 2009. PMID: 21089497 Review. Japanese. No abstract available.
-
DNA-RNA hybrid formation mediates RNAi-directed heterochromatin formation.Genes Cells. 2012 Mar;17(3):218-33. doi: 10.1111/j.1365-2443.2012.01583.x. Epub 2012 Jan 27. Genes Cells. 2012. PMID: 22280061
-
Epigenetics of imprinted long noncoding RNAs.Epigenetics. 2009 Jul 1;4(5):277-86. Epub 2009 Jul 10. Epigenetics. 2009. PMID: 19617707 Review.
-
New Insights into the Regulation of Heterochromatin.Trends Genet. 2016 May;32(5):284-294. doi: 10.1016/j.tig.2016.02.005. Epub 2016 Mar 20. Trends Genet. 2016. PMID: 27005444 Free PMC article. Review.
Cited by
-
RNA interference pathways in fungi: mechanisms and functions.Annu Rev Microbiol. 2012;66:305-23. doi: 10.1146/annurev-micro-092611-150138. Epub 2012 Jun 28. Annu Rev Microbiol. 2012. PMID: 22746336 Free PMC article. Review.
-
A SWI/SNF chromatin-remodeling complex acts in noncoding RNA-mediated transcriptional silencing.Mol Cell. 2013 Jan 24;49(2):298-309. doi: 10.1016/j.molcel.2012.11.011. Epub 2012 Dec 13. Mol Cell. 2013. PMID: 23246435 Free PMC article.
-
Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome.Genes Dev. 2009 Aug 15;23(16):1831-42. doi: 10.1101/gad.1811209. Genes Dev. 2009. PMID: 19684108 Free PMC article.
-
A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression.Genes Dev. 2014 Jul 1;28(13):1384-96. doi: 10.1101/gad.242990.114. Genes Dev. 2014. PMID: 24990962 Free PMC article. Review.
-
Elimination of a specific histone H3K14 acetyltransferase complex bypasses the RNAi pathway to regulate pericentric heterochromatin functions.Genes Dev. 2011 Feb 1;25(3):214-9. doi: 10.1101/gad.1993611. Genes Dev. 2011. PMID: 21289066 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

