Subcellular localization and functional analysis of the Arabidopsis GTPase RabE
- PMID: 19233904
- PMCID: PMC2663744
- DOI: 10.1104/pp.108.132092
Subcellular localization and functional analysis of the Arabidopsis GTPase RabE
Abstract
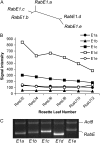
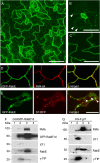
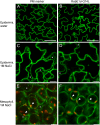
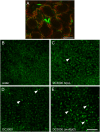
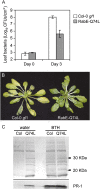
Membrane trafficking plays a fundamental role in eukaryotic cell biology. Of the numerous known or predicted protein components of the plant cell trafficking system, only a relatively small subset have been characterized with respect to their biological roles in plant growth, development, and response to stresses. In this study, we investigated the subcellular localization and function of an Arabidopsis (Arabidopsis thaliana) small GTPase belonging to the RabE family. RabE proteins are phylogenetically related to well-characterized regulators of polarized vesicle transport from the Golgi apparatus to the plasma membrane in animal and yeast cells. The RabE family of GTPases has also been proposed to be a putative host target of AvrPto, an effector protein produced by the plant pathogen Pseudomonas syringae, based on yeast two-hybrid analysis. We generated transgenic Arabidopsis plants that constitutively expressed one of the five RabE proteins (RabE1d) fused to green fluorescent protein (GFP). GFP-RabE1d and endogenous RabE proteins were found to be associated with the Golgi apparatus and the plasma membrane in Arabidopsis leaf cells. RabE down-regulation, due to cosuppression in transgenic plants, resulted in drastically altered leaf morphology and reduced plant size, providing experimental evidence for an important role of RabE GTPases in regulating plant growth. RabE down-regulation did not affect plant susceptibility to pathogenic P. syringae bacteria; conversely, expression of the constitutively active RabE1d-Q74L enhanced plant defenses, conferring resistance to P. syringae infection.
Figures



Similar articles
-
Arabidopsis actin-depolymerizing factor AtADF4 mediates defense signal transduction triggered by the Pseudomonas syringae effector AvrPphB.Plant Physiol. 2009 Jun;150(2):815-24. doi: 10.1104/pp.109.137604. Epub 2009 Apr 3. Plant Physiol. 2009. PMID: 19346440 Free PMC article.
-
Expression of antimicrobial peptides thanatin(S) in transgenic Arabidopsis enhanced resistance to phytopathogenic fungi and bacteria.Gene. 2013 Sep 15;527(1):235-42. doi: 10.1016/j.gene.2013.06.037. Epub 2013 Jun 29. Gene. 2013. PMID: 23820081
-
The role of Arabidopsis heterotrimeric G-protein subunits in MLO2 function and MAMP-triggered immunity.Mol Plant Microbe Interact. 2013 Sep;26(9):991-1003. doi: 10.1094/MPMI-03-13-0077-R. Mol Plant Microbe Interact. 2013. PMID: 23656333 Free PMC article.
-
Functional analysis of Arabidopsis WRKY25 transcription factor in plant defense against Pseudomonas syringae.BMC Plant Biol. 2007 Jan 10;7:2. doi: 10.1186/1471-2229-7-2. BMC Plant Biol. 2007. PMID: 17214894 Free PMC article.
-
More than meets the eye: knowns and unknowns of the trafficking of small secreted proteins in Arabidopsis.J Exp Bot. 2024 Jun 24;75(12):3713-3730. doi: 10.1093/jxb/erae172. J Exp Bot. 2024. PMID: 38693754 Review.
Cited by
-
A pathogen effector co-opts a host RabGAP protein to remodel pathogen interface and subvert defense-related secretion.Sci Adv. 2024 Oct 4;10(40):eado9516. doi: 10.1126/sciadv.ado9516. Epub 2024 Oct 4. Sci Adv. 2024. PMID: 39365859 Free PMC article.
-
The nonphototrophic hypocotyl 3 (NPH3) domain protein NRL5 is a trafficking-associated GTPase essential for drought resistance.Sci Adv. 2024 Aug 9;10(32):eado5429. doi: 10.1126/sciadv.ado5429. Epub 2024 Aug 9. Sci Adv. 2024. PMID: 39121213 Free PMC article.
-
Rab GTPases, tethers, and SNAREs work together to regulate Arabidopsis cell plate formation.Front Plant Sci. 2023 Feb 10;14:1120841. doi: 10.3389/fpls.2023.1120841. eCollection 2023. Front Plant Sci. 2023. PMID: 36844074 Free PMC article. Review.
-
Proteomic characterization of isolated Arabidopsis clathrin-coated vesicles reveals evolutionarily conserved and plant-specific components.Plant Cell. 2022 May 24;34(6):2150-2173. doi: 10.1093/plcell/koac071. Plant Cell. 2022. PMID: 35218346 Free PMC article.
-
Logistics of defense: The contribution of endomembranes to plant innate immunity.J Cell Biol. 2024 Jun 3;223(6):e202307066. doi: 10.1083/jcb.202307066. Epub 2024 Mar 29. J Cell Biol. 2024. PMID: 38551496 Free PMC article. Review.
References
-
- Alfano JR, Collmer A (2004) Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu Rev Phytopathol 42 385–414 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

