Analysis of PKR structure by small-angle scattering
- PMID: 19232355
- PMCID: PMC2663012
- DOI: 10.1016/j.jmb.2009.02.019
Analysis of PKR structure by small-angle scattering
Abstract
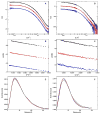
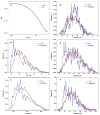
Protein kinase R (PKR) is a key component of the interferon antiviral defense pathway. Upon binding double-stranded RNA, PKR undergoes autophosphorylation reactions that activate the kinase. PKR contains an N-terminal double-stranded RNA binding domain, which consists of two tandem double-stranded RNA binding motifs, and a C-terminal kinase domain. We have used small-angle X-ray scattering and small-angle neutron scattering to define the conformation of latent PKR in solution. Guinier analysis indicates a radius of gyration of about 35 A. The p(r) distance distribution function exhibits a peak near 30 A, with a broad shoulder extending to longer distances. Good fits to the scattering data require models that incorporate multiple compact and extended conformations of the two interdomain linker regions. Thus, PKR belongs to the growing family of proteins that contain intrinsically unstructured regions. We propose that the flexible linkers may allow PKR to productively dimerize upon interaction with RNA activators that have diverse structures.
Figures
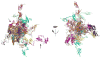
Similar articles
-
Specificity of the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR for double-stranded RNA: insights from thermodynamics and small-angle X-ray scattering.Biochemistry. 2012 Nov 20;51(46):9312-22. doi: 10.1021/bi300935p. Epub 2012 Nov 9. Biochemistry. 2012. PMID: 23140277 Free PMC article.
-
Recognition of viral RNA stem-loops by the tandem double-stranded RNA binding domains of PKR.RNA. 2013 Mar;19(3):333-44. doi: 10.1261/rna.035931.112. Epub 2013 Jan 17. RNA. 2013. PMID: 23329698 Free PMC article.
-
Role of the Interdomain Linker in RNA-Activated Protein Kinase Activation.Biochemistry. 2016 Jan 19;55(2):253-61. doi: 10.1021/acs.biochem.5b01171. Epub 2015 Dec 30. Biochemistry. 2016. PMID: 26678943 Free PMC article.
-
The search for a PKR code-differential regulation of protein kinase R activity by diverse RNA and protein regulators.RNA. 2019 May;25(5):539-556. doi: 10.1261/rna.070169.118. Epub 2019 Feb 15. RNA. 2019. PMID: 30770398 Free PMC article. Review.
-
Discriminating Self and Non-Self by RNA: Roles for RNA Structure, Misfolding, and Modification in Regulating the Innate Immune Sensor PKR.Acc Chem Res. 2016 Jun 21;49(6):1242-9. doi: 10.1021/acs.accounts.6b00151. Epub 2016 Jun 8. Acc Chem Res. 2016. PMID: 27269119 Free PMC article. Review.
Cited by
-
Analysis of monomeric and dimeric phosphorylated forms of protein kinase R.Biochemistry. 2010 Feb 16;49(6):1217-25. doi: 10.1021/bi901873p. Biochemistry. 2010. PMID: 20088595 Free PMC article.
-
Effect of interdomain dynamics on the structure determination of modular proteins by small-angle scattering.Eur Biophys J. 2010 Apr;39(5):769-80. doi: 10.1007/s00249-009-0549-3. Epub 2009 Oct 21. Eur Biophys J. 2010. PMID: 19844700
-
Structural Insights into High Density Lipoprotein: Old Models and New Facts.Front Pharmacol. 2016 Jan 12;6:318. doi: 10.3389/fphar.2015.00318. eCollection 2015. Front Pharmacol. 2016. PMID: 26793109 Free PMC article. Review.
-
The eIF2α kinases: their structures and functions.Cell Mol Life Sci. 2013 Oct;70(19):3493-511. doi: 10.1007/s00018-012-1252-6. Epub 2013 Jan 26. Cell Mol Life Sci. 2013. PMID: 23354059 Free PMC article. Review.
-
Specificity of the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR for double-stranded RNA: insights from thermodynamics and small-angle X-ray scattering.Biochemistry. 2012 Nov 20;51(46):9312-22. doi: 10.1021/bi300935p. Epub 2012 Nov 9. Biochemistry. 2012. PMID: 23140277 Free PMC article.
References
-
- Kaufman RJ. The double stranded RNA-activated protein kinase PKR. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2000. pp. 503–528.
-
- Toth AM, Zhang P, Das S, George CX, Samuel CE. Interferon action and the double-stranded RNA-dependent enzymes ADAR1 adenosine deaminase and PKR protein kinase. Prog Nucleic Acid Res Mol Biol. 2006;81:369–434. - PubMed
-
- Koromilas AE, Roy S, Barber GN, Katze MG, Sonenberg N. Malignant transformation by a mutant of the IFN-inducible dsRNA-dependent protein kinase. Science. 1992;257:1685–1689. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

