Relative contributions of the Candida albicans ABC transporters Cdr1p and Cdr2p to clinical azole resistance
- PMID: 19223631
- PMCID: PMC2663127
- DOI: 10.1128/AAC.00926-08
Relative contributions of the Candida albicans ABC transporters Cdr1p and Cdr2p to clinical azole resistance
Abstract
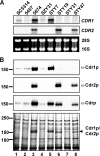
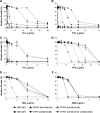
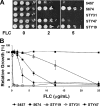
Candida albicans frequently develops resistance to treatment with azole drugs due to the acquisition of gain-of-function mutations in the transcription factor Tac1p. Tac1p hyperactivation in azole-resistant isolates results in the constitutive overexpression of several genes, including CDR1 and CDR2, which encode two homologous transporters of the ATP-binding cassette family. Functional studies of Cdr1p and Cdr2p have been carried out so far by heterologous expression in the budding yeast Saccharomyces cerevisiae and by gene deletion or overexpression in azole-sensitive C. albicans strains in which CDR1 expression is low and CDR2 expression is undetectable. Thus, the direct demonstration that CDR1 and CDR2 overexpression causes azole resistance in clinical strains is still lacking, as is our knowledge of the relative contribution of each transporter to clinical azole resistance. In the present study, we used the SAT1 flipper system to delete the CDR1 and CDR2 genes from clinical isolate 5674. This strain is resistant to several azole derivatives due to a strong hyperactive mutation in Tac1p and expresses high levels of Cdr1p and Cdr2p. We found that deleting CDR1 had a major effect, reducing resistance to fluconazole (FLC), ketoconazole (KTC), and itraconazole (ITC) by 6-, 4-, and 8-fold, respectively. Deleting CDR2 had a much weaker effect, reducing FLC or KTC resistance by 1.5-fold, and had no effect on ITC resistance. These results demonstrate that Cdr1p is a major determinant of azole resistance in strain 5674 and potentially in other clinical strains overexpressing Cdr1p and Cdr2p, while Cdr2p plays a more minor role.
Figures

Similar articles
-
ABC transporter Cdr1p contributes more than Cdr2p does to fluconazole efflux in fluconazole-resistant Candida albicans clinical isolates.Antimicrob Agents Chemother. 2008 Nov;52(11):3851-62. doi: 10.1128/AAC.00463-08. Epub 2008 Aug 18. Antimicrob Agents Chemother. 2008. PMID: 18710914 Free PMC article.
-
Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene.Microbiology (Reading). 1997 Feb;143 ( Pt 2):405-416. doi: 10.1099/00221287-143-2-405. Microbiology (Reading). 1997. PMID: 9043118
-
Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters.Antimicrob Agents Chemother. 1995 Nov;39(11):2378-86. doi: 10.1128/AAC.39.11.2378. Antimicrob Agents Chemother. 1995. PMID: 8585712 Free PMC article.
-
Antifungal drug resistance in pathogenic fungi.Med Mycol. 1998;36 Suppl 1:119-28. Med Mycol. 1998. PMID: 9988500 Review.
-
The TAC1 Gene in Candida albicans: Structure, Function, and Role in Azole Resistance: A Mini-Review.Microb Drug Resist. 2024 Jul;30(7):288-296. doi: 10.1089/mdr.2023.0334. Epub 2024 May 21. Microb Drug Resist. 2024. PMID: 38770776 Review.
Cited by
-
Engineering a Cysteine-Deficient Functional Candida albicans Cdr1 Molecule Reveals a Conserved Region at the Cytosolic Apex of ABCG Transporters Important for Correct Folding and Trafficking of Cdr1.mSphere. 2021 Feb 10;6(1):e01318-20. doi: 10.1128/mSphere.01318-20. mSphere. 2021. PMID: 33568458 Free PMC article.
-
Myriocin enhances the antifungal activity of fluconazole by blocking the membrane localization of the efflux pump Cdr1.Front Pharmacol. 2022 Dec 21;13:1101553. doi: 10.3389/fphar.2022.1101553. eCollection 2022. Front Pharmacol. 2022. PMID: 36618949 Free PMC article.
-
Azole susceptibility and transcriptome profiling in Candida albicans mitochondrial electron transport chain complex I mutants.Antimicrob Agents Chemother. 2013 Jan;57(1):532-42. doi: 10.1128/AAC.01520-12. Epub 2012 Nov 12. Antimicrob Agents Chemother. 2013. PMID: 23147730 Free PMC article.
-
An oxindole efflux inhibitor potentiates azoles and impairs virulence in the fungal pathogen Candida auris.Nat Commun. 2020 Dec 22;11(1):6429. doi: 10.1038/s41467-020-20183-3. Nat Commun. 2020. PMID: 33353950 Free PMC article.
-
Antifungal activity of the repurposed drug disulfiram against Cryptococcus neoformans.Front Pharmacol. 2024 Jan 11;14:1268649. doi: 10.3389/fphar.2023.1268649. eCollection 2023. Front Pharmacol. 2024. PMID: 38273827 Free PMC article.
References
-
- Akins, R. A. 2005. An update on antifungal targets and mechanisms of resistance in Candida albicans. Med. Mycol. 43:285-318. - PubMed
-
- Balzi, E., M. Wang, S. Leterme, D. L. Van, and A. Goffeau. 1994. PDR5, a novel yeast multidrug resistance conferring transporter controlled by the transcription regulator PDR1. J. Biol. Chem. 269:2206-2214. - PubMed
-
- Banerjee, D., N. Martin, S. Nandi, S. Shukla, A. Dominguez, G. Mukhopadhyay, and R. Prasad. 2007. A genome-wide steroid response study of the major human fungal pathogen Candida albicans. Mycopathologia 164:1-17. - PubMed
-
- Coste, A., V. Turner, F. Ischer, J. Morschhäuser, A. Forche, A. Selmecki, J. Berman, J. Bille, and D. Sanglard. 2006. A mutation in Tac1p, a transcription factor regulating CDR1 and CDR2, is coupled with loss of heterozygosity at chromosome 5 to mediate antifungal resistance in Candida albicans. Genetics 172:2139-2156. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

