Talin 2 is a large and complex gene encoding multiple transcripts and protein isoforms
- PMID: 19220457
- PMCID: PMC2702505
- DOI: 10.1111/j.1742-4658.2009.06893.x
Talin 2 is a large and complex gene encoding multiple transcripts and protein isoforms
Abstract
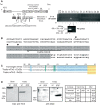
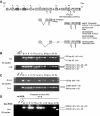
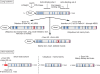
Talins are large adaptor proteins that link the integrin family of adhesion molecules to F-actin. In vertebrates, there are two talin genes. Talin 1 is essential for integrin-mediated cell adhesion; the role of talin 2 is unclear. Here we report a detailed analysis of mammalian talin 2. This reveals the existence of a previously unrecognized promoter associated with a CpG island, and separated from the first coding exon by numerous alternatively spliced noncoding exons spanning > 200 kb. Analysis of a mouse gene trap line shows that this promoter accounts for most of the talin 2 expression in adult tissues. We also demonstrate that testis and kidney express truncated talin 2 isoforms that lack the N-terminal half of the protein, and provide evidence for the developmentally regulated expression of the short testis-specific talin 2 isoform in elongating spermatids. Finally, we identify four tissue-specific alternative splicing events within the coding region of talin 2.
Figures



Similar articles
-
Evidence that talin alternative splice variants from Ciona intestinalis have different roles in cell adhesion.BMC Cell Biol. 2006 Dec 6;7:40. doi: 10.1186/1471-2121-7-40. BMC Cell Biol. 2006. PMID: 17150103 Free PMC article.
-
The tale of two talins - two isoforms to fine-tune integrin signalling.FEBS Lett. 2018 Jun;592(12):2108-2125. doi: 10.1002/1873-3468.13081. Epub 2018 May 18. FEBS Lett. 2018. PMID: 29723415 Free PMC article. Review.
-
Tissue-specific and ubiquitous promoters direct the expression of alternatively spliced transcripts from the calcitonin receptor gene.J Biol Chem. 2001 Jun 22;276(25):22663-74. doi: 10.1074/jbc.M007104200. Epub 2001 Apr 17. J Biol Chem. 2001. PMID: 11309373
-
Gene duplication and functional divergence during evolution of the cytoskeletal linker protein talin.Gene. 2005 Dec 5;362:141-52. doi: 10.1016/j.gene.2005.08.012. Epub 2005 Oct 10. Gene. 2005. PMID: 16216449
-
The human basonuclin 2 gene has the potential to generate nearly 90,000 mRNA isoforms encoding over 2000 different proteins.Genomics. 2007 Jan;89(1):44-58. doi: 10.1016/j.ygeno.2006.07.006. Epub 2006 Aug 30. Genomics. 2007. PMID: 16942855
Cited by
-
Endothelial PDGF-BB/PDGFR-β signaling promotes osteoarthritis by enhancing angiogenesis-dependent abnormal subchondral bone formation.Bone Res. 2022 Aug 29;10(1):58. doi: 10.1038/s41413-022-00229-6. Bone Res. 2022. PMID: 36031625 Free PMC article.
-
TLN1 contains a cancer-associated cassette exon that alters talin-1 mechanosensitivity.J Cell Biol. 2023 May 1;222(5):e202209010. doi: 10.1083/jcb.202209010. Epub 2023 Mar 6. J Cell Biol. 2023. PMID: 36880935 Free PMC article.
-
Talin1 regulates integrin turnover to promote embryonic epithelial morphogenesis.Mol Cell Biol. 2011 Aug;31(16):3366-77. doi: 10.1128/MCB.01403-10. Epub 2011 Jun 13. Mol Cell Biol. 2011. PMID: 21670148 Free PMC article.
-
The Mechanical Basis of Memory - the MeshCODE Theory.Front Mol Neurosci. 2021 Feb 25;14:592951. doi: 10.3389/fnmol.2021.592951. eCollection 2021. Front Mol Neurosci. 2021. PMID: 33716664 Free PMC article.
-
Both Talin-1 and Talin-2 correlate with malignancy potential of the human hepatocellular carcinoma MHCC-97 L cell.BMC Cancer. 2016 Jan 28;16:45. doi: 10.1186/s12885-016-2076-9. BMC Cancer. 2016. PMID: 26822056 Free PMC article.
References
-
- Critchley DR, Gingras AR. Talin at a glance. J Cell Sci. 2008;121:1345–1347. - PubMed
-
- Calderwood DA. Integrin activation. J Cell Sci. 2004;117:657–666. - PubMed
-
- Calderwood DA, Zent R, Grant R, Rees DJG, Hynes RO, Ginsberg MH. The talin head domain binds to integrin b subunit cytoplasmic tails and regulates integrin activation. J Biol Chem. 1999;274:28071–28704. - PubMed
-
- Lee HS, Bellin RM, Walker DL, Patel B, Powers P, Liu H, Garcia-Alvarez B, de Pereda JM, Liddington RC, Volkmann N, et al. Characterization of an actin-binding site within the talin FERM domain. J Mol Biol. 2004;343:771–784. - PubMed
-
- Loer B, Bauer R, Bornheim R, Grell J, Kremmer E, Kolanus W, Hoch M. The NHL-domain protein Wech is crucial for the integrin–cytoskeleton link. Nat Cell Biol. 2008;10:422–428. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

