Inhibition of SUV39H1 methyltransferase activity by DBC1
- PMID: 19218236
- PMCID: PMC2667723
- DOI: 10.1074/jbc.M900956200
Inhibition of SUV39H1 methyltransferase activity by DBC1
Abstract
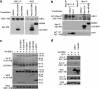
SUV39H1 is a histone H3K9-specific methyltransferase important for heterochromatin formation, regulation of gene expression, and induction of senescence in premalignant cells. SUV39H1 forms a complex with SirT1, and its activity is stimulated by SirT1 binding. Here we present evidence that the product of the DBC1 (deleted in breast cancer 1) gene disrupts the SUV39H1-SirT1 complex. Furthermore, DBC1 binds to the SUV39H1 catalytic domain and inhibits its ability to methylate histone H3 in vitro and in vivo. Knockdown of endogenous DBC1 increased the level of cellular H3K9 methylation. As expected, DBC1 also binds to SirT1 and inhibits the deacetylase activity of SirT1. These results identify DBC1 as a novel cellular inhibitor of SUV39H1 activity. DBC1 may be an important regulator of heterochromatin formation and genomic stability by disrupting the SUV39H1-SirT1 complex and inactivating both enzymes.
Figures


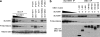

References
-
- Peters, A. H., O'Carroll, D., Scherthan, H., Mechtler, K., Sauer, S., Schofer, C., Weipoltshammer, K., Pagani, M., Lachner, M., Kohlmaier, A., Opravil, S., Doyle, M., Sibilia, M., and Jenuwein, T. (2001) Cell 107 323–337 - PubMed
-
- Nielsen, S. J., Schneider, R., Bauer, U. M., Bannister, A. J., Morrison, A., O'Carroll, D., Firestein, R., Cleary, M., Jenuwein, T., Herrera, R. E., and Kouzarides, T. (2001) Nature 412 561–565 - PubMed
-
- Braig, M., Lee, S., Loddenkemper, C., Rudolph, C., Peters, A. H., Schlegelberger, B., Stein, H., Dorken, B., Jenuwein, T., and Schmitt, C. A. (2005) Nature 436 660–665 - PubMed
-
- Imai, S., Armstrong, C. M., Kaeberlein, M., and Guarente, L. (2000) Nature 403 795–800 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

