A novel diagnostic target in the hepatitis C virus genome
- PMID: 19209955
- PMCID: PMC2637920
- DOI: 10.1371/journal.pmed.1000031
A novel diagnostic target in the hepatitis C virus genome
Abstract
Background: Detection and quantification of hepatitis C virus (HCV) RNA is integral to diagnostic and therapeutic regimens. All molecular assays target the viral 5'-noncoding region (5'-NCR), and all show genotype-dependent variation of sensitivities and viral load results. Non-western HCV genotypes have been under-represented in evaluation studies. An alternative diagnostic target region within the HCV genome could facilitate a new generation of assays.
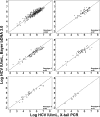
Methods and findings: In this study we determined by de novo sequencing that the 3'-X-tail element, characterized significantly later than the rest of the genome, is highly conserved across genotypes. To prove its clinical utility as a molecular diagnostic target, a prototype qualitative and quantitative test was developed and evaluated multicentrically on a large and complete panel of 725 clinical plasma samples, covering HCV genotypes 1-6, from four continents (Germany, UK, Brazil, South Africa, Singapore). To our knowledge, this is the most diversified and comprehensive panel of clinical and genotype specimens used in HCV nucleic acid testing (NAT) validation to date. The lower limit of detection (LOD) was 18.4 IU/ml (95% confidence interval, 15.3-24.1 IU/ml), suggesting applicability in donor blood screening. The upper LOD exceeded 10(-9) IU/ml, facilitating viral load monitoring within a wide dynamic range. In 598 genotyped samples, quantified by Bayer VERSANT 3.0 branched DNA (bDNA), X-tail-based viral loads were highly concordant with bDNA for all genotypes. Correlation coefficients between bDNA and X-tail NAT, for genotypes 1-6, were: 0.92, 0.85, 0.95, 0.91, 0.95, and 0.96, respectively; X-tail-based viral loads deviated by more than 0.5 log10 from 5'-NCR-based viral loads in only 12% of samples (maximum deviation, 0.85 log10). The successful introduction of X-tail NAT in a Brazilian laboratory confirmed the practical stability and robustness of the X-tail-based protocol. The assay was implemented at low reaction costs (US$8.70 per sample), short turnover times (2.5 h for up to 96 samples), and without technical difficulties.
Conclusion: This study indicates a way to fundamentally improve HCV viral load monitoring and infection screening. Our prototype assay can serve as a template for a new generation of viral load assays. Additionally, to our knowledge this study provides the first open protocol to permit industry-grade HCV detection and quantification in resource-limited settings.
Conflict of interest statement
Figures


Comment in
-
What are the prospects for controlling hepatitis C?PLoS Med. 2009 Jun 16;6(6):e1000096. doi: 10.1371/journal.pmed.1000096. Epub 2009 Jun 16. PLoS Med. 2009. PMID: 19529757 Free PMC article. No abstract available.
Similar articles
-
Multilaboratory comparison of hepatitis C virus viral load assays.J Clin Microbiol. 2006 May;44(5):1726-32. doi: 10.1128/JCM.44.5.1726-1732.2006. J Clin Microbiol. 2006. PMID: 16672399 Free PMC article.
-
Comparative evaluation of the VERSANT HCV RNA 3.0, QUANTIPLEX HCV RNA 2.0, and COBAS AMPLICOR HCV MONITOR version 2.0 Assays for quantification of hepatitis C virus RNA in serum.J Clin Microbiol. 2002 Feb;40(2):495-500. doi: 10.1128/JCM.40.2.495-500.2002. J Clin Microbiol. 2002. PMID: 11825962 Free PMC article.
-
Successful HCV genotyping of previously failed and low viral load specimens using an HCV RNA qualitative assay based on transcription-mediated amplification in conjunction with the line probe assay.J Clin Virol. 2003 Sep;28(1):14-26. doi: 10.1016/s1386-6532(02)00234-2. J Clin Virol. 2003. PMID: 12927747
-
Hepatitis C Core Antigen Testing for Diagnosis of Hepatitis C Virus Infection: A Systematic Review and Meta-analysis.Ann Intern Med. 2016 Sep 6;165(5):345-55. doi: 10.7326/M16-0065. Epub 2016 Jun 21. Ann Intern Med. 2016. PMID: 27322622 Free PMC article. Review.
-
Diagnostic Accuracy of Point-of-Care HCV Viral Load Assays for HCV Diagnosis: A Systematic Review and Meta-Analysis.Diagnostics (Basel). 2022 May 18;12(5):1255. doi: 10.3390/diagnostics12051255. Diagnostics (Basel). 2022. PMID: 35626411 Free PMC article. Review.
Cited by
-
Differential Infection Patterns and Recent Evolutionary Origins of Equine Hepaciviruses in Donkeys.J Virol. 2016 Dec 16;91(1):e01711-16. doi: 10.1128/JVI.01711-16. Print 2017 Jan 1. J Virol. 2016. PMID: 27795428 Free PMC article.
-
Bile acids specifically increase hepatitis C virus RNA-replication.PLoS One. 2012;7(4):e36029. doi: 10.1371/journal.pone.0036029. Epub 2012 Apr 25. PLoS One. 2012. PMID: 22558311 Free PMC article.
-
A novel hepatitis C virus genotyping method based on liquid microarray.PLoS One. 2010 Sep 20;5(9):e12822. doi: 10.1371/journal.pone.0012822. PLoS One. 2010. PMID: 20862224 Free PMC article.
-
External Quality Assessment for Zika Virus Molecular Diagnostic Testing, Brazil.Emerg Infect Dis. 2018 May;24(5):888-92. doi: 10.3201/eid2405.171747. Epub 2018 May 17. Emerg Infect Dis. 2018. PMID: 29470164 Free PMC article.
-
Molecular detection of human rhinoviruses in respiratory samples: a comparison of Taqman probe-, SYBR green I- and BOXTO-based real-time PCR assays.Virol J. 2014 Feb 18;11:31. doi: 10.1186/1743-422X-11-31. Virol J. 2014. PMID: 24548758 Free PMC article.
References
-
- Poynard T, Yuen MF, Ratziu V, Lai CL. Viral hepatitis C. Lancet. 2003;362:2095–2100. - PubMed
-
- Perz JF, Alter MJ. The coming wave of HCV-related liver disease: dilemmas and challenges. J Hepatol. 2006;44:441–443. - PubMed
-
- Anonymous. Global surveillance and control of hepatitis C. Report of a WHO Consultation organized in collaboration with the Viral Hepatitis Prevention Board, Antwerp, Belgium. J Viral Hepat. 1999;6:35–47. - PubMed
-
- Simmonds P, Bukh J, Combet C, Deleage G, Enomoto N, et al. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42:962–973. - PubMed
-
- Berg T, Sarrazin C, Herrmann E, Hinrichsen H, Gerlach T, et al. Prediction of treatment outcome in patients with chronic hepatitis C: significance of baseline parameters and viral dynamics during therapy. Hepatology. 2003;37:600–609. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

