Reconstitution and dissection of the 600-kDa Srv2/CAP complex: roles for oligomerization and cofilin-actin binding in driving actin turnover
- PMID: 19201756
- PMCID: PMC2667778
- DOI: 10.1074/jbc.M808760200
Reconstitution and dissection of the 600-kDa Srv2/CAP complex: roles for oligomerization and cofilin-actin binding in driving actin turnover
Abstract
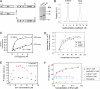
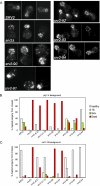
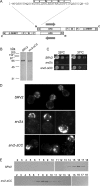
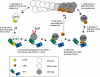
Srv2/cyclase-associated protein is expressed in virtually all plant, animal, and fungal organisms and has a conserved role in promoting actin depolymerizing factor/cofilin-mediated actin turnover. This is achieved by the abilities of Srv2 to recycle cofilin from ADP-actin monomers and to promote nucleotide exchange (ATP for ADP) on actin monomers. Despite this important and universal role in facilitating actin turnover, the mechanism underlying Srv2 function has remained elusive. Previous studies have demonstrated a critical functional role for the G-actin-binding C-terminal half of Srv2. Here we describe an equally important role in vivo for the N-terminal half of Srv2 in driving actin turnover. We pinpoint this activity to a conserved patch of surface residues on the N-terminal dimeric helical folded domain of Srv2, and we show that this functional site interacts with cofilin-actin complexes. Furthermore, we show that this site is essential for Srv2 acceleration of cofilin-mediated actin turnover in vitro. A cognate Srv2-binding site is identified on a conserved surface of cofilin, suggesting that this function likely extends to other organisms. In addition, our analyses reveal that higher order oligomerization of Srv2 depends on its N-terminal predicted coiled coil domain and that oligomerization optimizes Srv2 function in vitro and in vivo. Based on these data, we present a revised model for the mechanism by which Srv2 promotes actin turnover, in which coordinated activities of its N- and C-terminal halves catalyze sequential steps in recycling cofilin and actin monomers.
Figures



Similar articles
-
The role of cyclase-associated protein in regulating actin filament dynamics - more than a monomer-sequestration factor.J Cell Sci. 2013 Aug 1;126(Pt 15):3249-58. doi: 10.1242/jcs.128231. J Cell Sci. 2013. PMID: 23908377 Free PMC article. Review.
-
Structure and mechanism of mouse cyclase-associated protein (CAP1) in regulating actin dynamics.J Biol Chem. 2014 Oct 31;289(44):30732-30742. doi: 10.1074/jbc.M114.601765. Epub 2014 Sep 16. J Biol Chem. 2014. PMID: 25228691 Free PMC article.
-
A high-affinity interaction with ADP-actin monomers underlies the mechanism and in vivo function of Srv2/cyclase-associated protein.Mol Biol Cell. 2004 Nov;15(11):5158-71. doi: 10.1091/mbc.e04-06-0444. Epub 2004 Sep 8. Mol Biol Cell. 2004. PMID: 15356265 Free PMC article.
-
Coordinated regulation of actin filament turnover by a high-molecular-weight Srv2/CAP complex, cofilin, profilin, and Aip1.Curr Biol. 2003 Dec 16;13(24):2159-69. doi: 10.1016/j.cub.2003.11.051. Curr Biol. 2003. PMID: 14680631
-
WH2 domain: a small, versatile adapter for actin monomers.FEBS Lett. 2002 Feb 20;513(1):92-7. doi: 10.1016/s0014-5793(01)03242-2. FEBS Lett. 2002. PMID: 11911886 Review.
Cited by
-
Mechanism of CAP1-mediated apical actin polymerization in pollen tubes.Proc Natl Acad Sci U S A. 2019 Jun 11;116(24):12084-12093. doi: 10.1073/pnas.1821639116. Epub 2019 May 23. Proc Natl Acad Sci U S A. 2019. PMID: 31123151 Free PMC article.
-
Autonomous and in trans functions for the two halves of Srv2/CAP in promoting actin turnover.Cytoskeleton (Hoboken). 2014 Jun;71(6):351-360. doi: 10.1002/cm.21170. Epub 2014 Apr 25. Cytoskeleton (Hoboken). 2014. PMID: 24616256 Free PMC article.
-
Phosphorylation of the cytoskeletal protein CAP1 controls its association with cofilin and actin.J Cell Sci. 2014 Dec 1;127(Pt 23):5052-65. doi: 10.1242/jcs.156059. Epub 2014 Oct 14. J Cell Sci. 2014. PMID: 25315833 Free PMC article.
-
The role of cyclase-associated protein in regulating actin filament dynamics - more than a monomer-sequestration factor.J Cell Sci. 2013 Aug 1;126(Pt 15):3249-58. doi: 10.1242/jcs.128231. J Cell Sci. 2013. PMID: 23908377 Free PMC article. Review.
-
Dynamic remodeling of actin networks by cyclase-associated protein and CAP-Abp1 complexes.Curr Biol. 2023 Oct 23;33(20):4484-4495.e5. doi: 10.1016/j.cub.2023.09.032. Epub 2023 Oct 4. Curr Biol. 2023. PMID: 37797614 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

