Genome-wide analysis of factors affecting transcription elongation and DNA repair: a new role for PAF and Ccr4-not in transcription-coupled repair
- PMID: 19197357
- PMCID: PMC2629578
- DOI: 10.1371/journal.pgen.1000364
Genome-wide analysis of factors affecting transcription elongation and DNA repair: a new role for PAF and Ccr4-not in transcription-coupled repair
Abstract
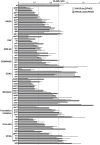
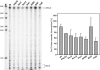
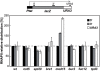
RNA polymerases frequently deal with a number of obstacles during transcription elongation that need to be removed for transcription resumption. One important type of hindrance consists of DNA lesions, which are removed by transcription-coupled repair (TC-NER), a specific sub-pathway of nucleotide excision repair. To improve our knowledge of transcription elongation and its coupling to TC-NER, we used the yeast library of non-essential knock-out mutations to screen for genes conferring resistance to the transcription-elongation inhibitor mycophenolic acid and the DNA-damaging agent 4-nitroquinoline-N-oxide. Our data provide evidence that subunits of the SAGA and Ccr4-Not complexes, Mediator, Bre1, Bur2, and Fun12 affect transcription elongation to different extents. Given the dependency of TC-NER on RNA Polymerase II transcription and the fact that the few proteins known to be involved in TC-NER are related to transcription, we performed an in-depth TC-NER analysis of a selection of mutants. We found that mutants of the PAF and Ccr4-Not complexes are impaired in TC-NER. This study provides evidence that PAF and Ccr4-Not are required for efficient TC-NER in yeast, unraveling a novel function for these transcription complexes and opening new perspectives for the understanding of TC-NER and its functional interconnection with transcription elongation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





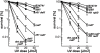
Similar articles
-
Evidence that Moderate Eviction of Spt5 and Promotion of Error-Free Transcriptional Bypass by Rad26 Facilitates Transcription Coupled Nucleotide Excision Repair.J Mol Biol. 2019 Mar 29;431(7):1322-1338. doi: 10.1016/j.jmb.2019.02.010. Epub 2019 Feb 18. J Mol Biol. 2019. PMID: 30790631
-
Transcription coupled nucleotide excision repair in the yeast Saccharomyces cerevisiae: The ambiguous role of Rad26.DNA Repair (Amst). 2015 Dec;36:43-48. doi: 10.1016/j.dnarep.2015.09.006. Epub 2015 Sep 10. DNA Repair (Amst). 2015. PMID: 26429063 Review.
-
Genome-wide role of Rad26 in promoting transcription-coupled nucleotide excision repair in yeast chromatin.Proc Natl Acad Sci U S A. 2020 Aug 4;117(31):18608-18616. doi: 10.1073/pnas.2003868117. Epub 2020 Jul 20. Proc Natl Acad Sci U S A. 2020. PMID: 32690696 Free PMC article.
-
Nucleotide excision repair and the 26S proteasome function together to promote trinucleotide repeat expansions.DNA Repair (Amst). 2014 Jan;13:42-9. doi: 10.1016/j.dnarep.2013.11.004. Epub 2013 Dec 17. DNA Repair (Amst). 2014. PMID: 24359926
-
The role of Cockayne syndrome group A (CSA) protein in transcription-coupled nucleotide excision repair.Mech Ageing Dev. 2013 May-Jun;134(5-6):196-201. doi: 10.1016/j.mad.2013.03.008. Epub 2013 Apr 6. Mech Ageing Dev. 2013. PMID: 23571135 Review.
Cited by
-
Ccr4-Not complex: the control freak of eukaryotic cells.Crit Rev Biochem Mol Biol. 2012 Jul-Aug;47(4):315-33. doi: 10.3109/10409238.2012.667214. Epub 2012 Mar 15. Crit Rev Biochem Mol Biol. 2012. PMID: 22416820 Free PMC article. Review.
-
Elf1 promotes transcription-coupled repair in yeast by using its C-terminal domain to bind TFIIH.Nat Commun. 2024 Jul 23;15(1):6223. doi: 10.1038/s41467-024-50539-y. Nat Commun. 2024. PMID: 39043658 Free PMC article.
-
The cellular roles of Ccr4-NOT in model and pathogenic fungi-implications for fungal virulence.Front Genet. 2013 Dec 20;4:302. doi: 10.3389/fgene.2013.00302. eCollection 2013. Front Genet. 2013. PMID: 24391665 Free PMC article.
-
Cleavage factor I links transcription termination to DNA damage response and genome integrity maintenance in Saccharomyces cerevisiae.PLoS Genet. 2014 Mar 6;10(3):e1004203. doi: 10.1371/journal.pgen.1004203. eCollection 2014 Mar. PLoS Genet. 2014. PMID: 24603480 Free PMC article.
-
Proteomic Analysis of the Mediator Complex Interactome in Saccharomyces cerevisiae.Sci Rep. 2017 Feb 27;7:43584. doi: 10.1038/srep43584. Sci Rep. 2017. PMID: 28240253 Free PMC article.
References
-
- Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–567. - PubMed
-
- Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 2005;17:251–256. - PubMed
-
- Rosonina E, Kaneko S, Manley JL. Terminating the transcript: breaking up is hard to do. Genes Dev. 2006;20:1050–1056. - PubMed
-
- Svejstrup JQ. Contending with transcriptional arrest during RNAPII transcript elongation. Trends Biochem Sci. 2007;32:165–171. - PubMed
-
- Mellon I. Transcription-coupled repair: a complex affair. Mutat Res. 2005;577:155–161. - PubMed

