Human cytomegalovirus glycoprotein B is required for virus entry and cell-to-cell spread but not for virion attachment, assembly, or egress
- PMID: 19193805
- PMCID: PMC2663263
- DOI: 10.1128/JVI.01251-08
Human cytomegalovirus glycoprotein B is required for virus entry and cell-to-cell spread but not for virion attachment, assembly, or egress
Abstract
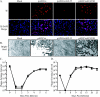
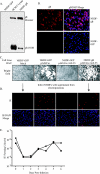
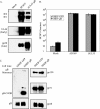
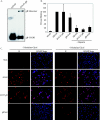
Glycoprotein B (gB) homologs are conserved throughout the family Herpesviridae and appear to serve essential, universal functions, as well as specific functions unique to a particular herpesvirus. Genetic analysis is a powerful tool to analyze protein function, and while it has been possible to generate virus mutants, complementation of essential virus knockouts has been problematic. Human cytomegalovirus (HCMV) gB (UL55) plays an essential role in the replication cycle of the virus. To define the function(s) of gB in HCMV infection, the BAC system was used to generate a recombinant virus in which the UL55 gene was replaced with galK (pAD/CreDeltaUL55). UL55 deletions in the viral genome have been made before, demonstrating that UL55 is an essential gene. However, without being able to successfully complement the genetic defect, a phenotypic analysis of the mutant virus was impossible. We generated fibroblasts expressing HCMV gB that complement pAD/CreDeltaUL55 and produce infectious virions lacking the UL55 gene but containing wild-type gB on the virion surface (DeltaUL55-gB HCMV). This is the first successful complementation of an HCMV mutant with a glycoprotein deleted. To characterize DeltaUL55 infection in the absence of gB, noncomplementing cells were infected with DeltaUL55-gB virus. All stages of gene expression were detected, and significant amounts of DNase-resistant viral DNA genomes, representing whole intact virions, were released into the infected cell supernatant. Gradient purification of these virions revealed they lacked gB but contained other viral structural proteins. The gB-null virions were able to attach to the cell surface similarly to wild-type gB-containing virions but were defective in virus entry and cell-to-cell spread. Glycoprotein B-null virions do, however, contain infectious DNA, as IE gene expression can be detected in fibroblasts following treatment of attached gB-null virions with a membrane fusion agent, polyethylene glycol. Taken together, our results indicate that gB is required for virus entry and cell-to-cell spread of the virus. However, HCMV gB is not absolutely required for virus attachment or assembly and egress from infected cells.
Figures


Similar articles
-
Human cytomegalovirus (HCMV) glycoprotein gB promotes virus entry in trans acting as the viral fusion protein rather than as a receptor-binding protein.mBio. 2013 Jun 4;4(3):e00332-13. doi: 10.1128/mBio.00332-13. mBio. 2013. PMID: 23736286 Free PMC article.
-
Loss of the Human Cytomegalovirus US16 Protein Abrogates Virus Entry into Endothelial and Epithelial Cells by Reducing the Virion Content of the Pentamer.J Virol. 2017 May 12;91(11):e00205-17. doi: 10.1128/JVI.00205-17. Print 2017 Jun 1. J Virol. 2017. PMID: 28331097 Free PMC article.
-
Human Cytomegalovirus gH/gL/gO Promotes the Fusion Step of Entry into All Cell Types, whereas gH/gL/UL128-131 Broadens Virus Tropism through a Distinct Mechanism.J Virol. 2015 Sep;89(17):8999-9009. doi: 10.1128/JVI.01325-15. Epub 2015 Jun 17. J Virol. 2015. PMID: 26085146 Free PMC article.
-
Tegument proteins of human cytomegalovirus.Microbiol Mol Biol Rev. 2008 Jun;72(2):249-65, table of contents. doi: 10.1128/MMBR.00040-07. Microbiol Mol Biol Rev. 2008. PMID: 18535146 Free PMC article. Review.
-
The unconventional way out-Egress of HCMV through multiviral bodies.Mol Microbiol. 2022 Jun;117(6):1317-1323. doi: 10.1111/mmi.14946. Epub 2022 Jun 6. Mol Microbiol. 2022. PMID: 35607767 Review.
Cited by
-
Identification of adipocyte plasma membrane-associated protein as a novel modulator of human cytomegalovirus infection.PLoS Pathog. 2019 Jul 29;15(7):e1007914. doi: 10.1371/journal.ppat.1007914. eCollection 2019 Jul. PLoS Pathog. 2019. PMID: 31356650 Free PMC article.
-
Human Cytomegalovirus Primary Infection and Reactivation: Insights From Virion-Carried Molecules.Front Microbiol. 2020 Jul 14;11:1511. doi: 10.3389/fmicb.2020.01511. eCollection 2020. Front Microbiol. 2020. PMID: 32765441 Free PMC article. Review.
-
Principles for studying in vivo attenuation of virus mutants: defining the role of the cytomegalovirus gH/gL/gO complex as a paradigm.Med Microbiol Immunol. 2015 Jun;204(3):295-305. doi: 10.1007/s00430-015-0405-2. Epub 2015 Mar 18. Med Microbiol Immunol. 2015. PMID: 25782576 Review.
-
Induction of Filopodia During Cytomegalovirus Entry Into Human Iris Stromal Cells.Front Microbiol. 2022 Apr 5;13:834927. doi: 10.3389/fmicb.2022.834927. eCollection 2022. Front Microbiol. 2022. PMID: 35450284 Free PMC article.
-
The human cytomegalovirus decathlon: Ten critical replication events provide opportunities for restriction.Front Cell Dev Biol. 2022 Nov 25;10:1053139. doi: 10.3389/fcell.2022.1053139. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36506089 Free PMC article. Review.
References
-
- Alford, C. A. and W. J. Britt. 1993. Cytomegalovirus, p. 227-255. In B. Roizman, R. J. Whitley, and C. Lopez (ed.), Human herpesviruses. Raven Press, New York, NY.
-
- Boehme, K. W., and T. Compton. 2006. Virus entry and activation of innate immunity, p. 111-130. In M. J. Reddehase (ed.), Cytomegaloviruses: molecular biology and immunology. Caister, Norfolk, United Kingdom.
-
- Boehme, K. W., M. Guerrero, and T. Compton. 2006. Human cytomegalovirus envelope glycoproteins B and H are necessary for TLR2 activation in permissive cells. J. Immunol. 1777094-7102. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

