Cryptic variation in the human mutation rate
- PMID: 19192947
- PMCID: PMC2634788
- DOI: 10.1371/journal.pbio.1000027
Cryptic variation in the human mutation rate
Abstract
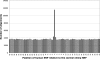
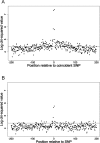
The mutation rate is known to vary between adjacent sites within the human genome as a consequence of context, the most well-studied example being the influence of CpG dinucelotides. We investigated whether there is additional variation by testing whether there is an excess of sites at which both humans and chimpanzees have a single-nucleotide polymorphism (SNP). We found a highly significant excess of such sites, and we demonstrated that this excess is not due to neighbouring nucleotide effects, ancestral polymorphism, or natural selection. We therefore infer that there is cryptic variation in the mutation rate. However, although this variation in the mutation rate is not associated with the adjacent nucleotides, we show that there are highly nonrandom patterns of nucleotides that extend approximately 80 base pairs on either side of sites with coincident SNPs, suggesting that there are extensive and complex context effects. Finally, we estimate the level of variation needed to produce the excess of coincident SNPs and show that there is a similar, or higher, level of variation in the mutation rate associated with this cryptic process than there is associated with adjacent nucleotides, including the CpG effect. We conclude that there is substantial variation in the mutation that has, until now, been hidden from view.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
Similar articles
-
The genomic distribution and local context of coincident SNPs in human and chimpanzee.Genome Biol Evol. 2010;2:547-57. doi: 10.1093/gbe/evq039. Epub 2010 Jul 8. Genome Biol Evol. 2010. PMID: 20675616 Free PMC article.
-
Strand bias in complementary single-nucleotide polymorphisms of transcribed human sequences: evidence for functional effects of synonymous polymorphisms.BMC Genomics. 2006 Aug 17;7:213. doi: 10.1186/1471-2164-7-213. BMC Genomics. 2006. PMID: 16916449 Free PMC article.
-
Effect of the assignment of ancestral CpG state on the estimation of nucleotide substitution rates in mammals.BMC Evol Biol. 2008 Sep 30;8:265. doi: 10.1186/1471-2148-8-265. BMC Evol Biol. 2008. PMID: 18826599 Free PMC article.
-
[Chimpanzee genome sequencing and comparative analysis with the human genome].Tanpakushitsu Kakusan Koso. 2006 Feb;51(2):178-87. Tanpakushitsu Kakusan Koso. 2006. PMID: 16457209 Review. Japanese. No abstract available.
-
Variation in the mutation rate across mammalian genomes.Nat Rev Genet. 2011 Oct 4;12(11):756-66. doi: 10.1038/nrg3098. Nat Rev Genet. 2011. PMID: 21969038 Review.
Cited by
-
A new test suggests hundreds of amino acid polymorphisms in humans are subject to balancing selection.PLoS Biol. 2022 Jun 2;20(6):e3001645. doi: 10.1371/journal.pbio.3001645. eCollection 2022 Jun. PLoS Biol. 2022. PMID: 35653351 Free PMC article.
-
A-to-I RNA editing contributes to the persistence of predicted damaging mutations in populations.Genome Res. 2019 Nov;29(11):1766-1776. doi: 10.1101/gr.246033.118. Epub 2019 Sep 12. Genome Res. 2019. PMID: 31515285 Free PMC article.
-
Comparative Investigation of Coincident Single Nucleotide Polymorphisms Underlying Avian Influenza Viruses in Chickens and Ducks.Biology (Basel). 2023 Jul 7;12(7):969. doi: 10.3390/biology12070969. Biology (Basel). 2023. PMID: 37508399 Free PMC article.
-
Identification of deleterious mutations within three human genomes.Genome Res. 2009 Sep;19(9):1553-61. doi: 10.1101/gr.092619.109. Epub 2009 Jul 14. Genome Res. 2009. PMID: 19602639 Free PMC article.
-
Quantifying Influences on Intragenomic Mutation Rate.G3 (Bethesda). 2020 Aug 5;10(8):2641-2652. doi: 10.1534/g3.120.401335. G3 (Bethesda). 2020. PMID: 32527747 Free PMC article.
References
-
- Miyata T, Hayashida H, Kuma K, Mitsuyasu K, Yasunaga T. Male-driven molecular evolution: a model and nucleotide sequence analysis. Cold Spring Harb Symp Quant Biol. 1987;52:863–867. - PubMed
-
- Li WH, Yi S, Makova K. Male-driven evolution. Curr Opin Genet Dev. 2002;12:650–656. - PubMed
-
- Lercher MJ, Williams EJ, Hurst LD. Local similarity in evolutionary rates extends over whole chromosomes in human-rodent and mouse-rat comparisons: implications for understanding the mechanistic basis of the male mutation bias. Mol Biol Evol. 2001;18:2032–2039. - PubMed
-
- Matassi G, Sharp PM, Gautier C. Chromosomal location effects on gene sequence evolution in mammals. Curr Biol. 1999;9:786–791. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

