Multiple SET methyltransferases are required to maintain normal heterochromatin domains in the genome of Drosophila melanogaster
- PMID: 19189944
- PMCID: PMC2666501
- DOI: 10.1534/genetics.108.100271
Multiple SET methyltransferases are required to maintain normal heterochromatin domains in the genome of Drosophila melanogaster
Abstract
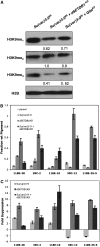
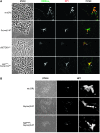
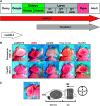
Methylation of histone H3 lysine 9 (H3K9) is a key feature of silent chromatin and plays an important role in stabilizing the interaction of heterochromatin protein 1 (HP1) with chromatin. Genomes of metazoans such as the fruit fly Drosophila melanogaster generally encode three types of H3K9-specific SET domain methyltransferases that contribute to chromatin homeostasis during the life cycle of the organism. SU(VAR)3-9, dG9a, and dSETDB1 all function in the generation of wild-type H3K9 methylation levels in the Drosophila genome. Two of these enzymes, dSETDB1 and SU(VAR)3-9, govern heterochromatin formation in distinct but overlapping patterns across the genome. H3K9 methylation in the small, heterochromatic fourth chromosome of D. melanogaster is governed mainly by dSETDB1, whereas dSETDB1 and SU(VAR)3-9 function in concert to methylate H3K9 in the pericentric heterochromatin of all chromosomes, with dG9a having little impact in these domains, as shown by monitoring position effect variegation. To understand how these distinct heterochromatin compartments may be differentiated, we examined the developmental timing of dSETDB1 function using a knockdown strategy. dSETDB1 acts to maintain heterochromatin during metamorphosis, at a later stage in development than the reported action of SU(VAR)3-9. Surprisingly, depletion of both of these enzymes has less deleterious effect than depletion of one. These results imply that dSETDB1 acts as a heterochromatin maintenance factor that may be required for the persistence of earlier developmental events normally governed by SU(VAR)3-9. In addition, the genetic interactions between dSETDB1 and Su(var)3-9 mutations emphasize the importance of maintaining the activities of these histone methyltransferases in balance for normal genome function.
Figures



Similar articles
-
Epigenetic regulation of the Drosophila chromosome 4 by the histone H3K9 methyltransferase dSETDB1.Proc Natl Acad Sci U S A. 2007 Jul 31;104(31):12691-6. doi: 10.1073/pnas.0705534104. Epub 2007 Jul 25. Proc Natl Acad Sci U S A. 2007. PMID: 17652514 Free PMC article.
-
Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing.EMBO J. 2002 Mar 1;21(5):1121-31. doi: 10.1093/emboj/21.5.1121. EMBO J. 2002. PMID: 11867540 Free PMC article.
-
dSETDB1 and SU(VAR)3-9 sequentially function during germline-stem cell differentiation in Drosophila melanogaster.PLoS One. 2008 May 21;3(5):e2234. doi: 10.1371/journal.pone.0002234. PLoS One. 2008. PMID: 18493619 Free PMC article.
-
Histone modification and the control of heterochromatic gene silencing in Drosophila.Chromosome Res. 2006;14(4):377-92. doi: 10.1007/s10577-006-1066-1. Chromosome Res. 2006. PMID: 16821134 Review.
-
SU(VAR)3-9 is a conserved key function in heterochromatic gene silencing.Genetica. 2003 Mar;117(2-3):149-58. doi: 10.1023/a:1022923508198. Genetica. 2003. PMID: 12723694 Review.
Cited by
-
HP1a, Su(var)3-9, SETDB1 and POF stimulate or repress gene expression depending on genomic position, gene length and expression pattern in Drosophila melanogaster.Nucleic Acids Res. 2013 Apr;41(8):4481-94. doi: 10.1093/nar/gkt158. Epub 2013 Mar 9. Nucleic Acids Res. 2013. PMID: 23476027 Free PMC article.
-
Epigenetic regulation of oogenesis and germ stem cell maintenance by the Drosophila histone methyltransferase Eggless/dSetDB1.Dev Biol. 2014 Apr 15;388(2):181-91. doi: 10.1016/j.ydbio.2014.01.014. Epub 2014 Jan 28. Dev Biol. 2014. PMID: 24485852 Free PMC article.
-
Mechanistic insights into KDM4A driven genomic instability.Biochem Soc Trans. 2021 Feb 26;49(1):93-105. doi: 10.1042/BST20191219. Biochem Soc Trans. 2021. PMID: 33492339 Free PMC article. Review.
-
Trans-inactivation: Repression in a wrong place.Fly (Austin). 2017 Apr 3;11(2):96-103. doi: 10.1080/19336934.2016.1225634. Epub 2016 Aug 19. Fly (Austin). 2017. PMID: 27540893 Free PMC article.
-
The impact of genetic background and cell lineage on the level and pattern of gene expression in position effect variegation.Epigenetics Chromatin. 2019 Nov 13;12(1):70. doi: 10.1186/s13072-019-0314-5. Epigenetics Chromatin. 2019. PMID: 31722719 Free PMC article.
References
-
- Benjamini, Y., and Y. Hochberg, 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57 289–300.
-
- Clough, E., W. Moon, S. Wang, K. Smith and T. Hazelrigg, 2007. Histone methylation is required for oogenesis in Drosophila. Development 134 157–165. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

