Chromosomal location targets different MYC family gene members for oncogenic translocations
- PMID: 19174520
- PMCID: PMC2650145
- DOI: 10.1073/pnas.0812763106
Chromosomal location targets different MYC family gene members for oncogenic translocations
Abstract
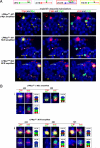
The MYC family of cellular oncogenes includes c-Myc, N-myc, and L-myc, which encode transcriptional regulators involved in the control of cell proliferation and death. Accordingly, these genes become aberrantly activated and expressed in specific types of cancers. For example, c-Myc translocations occur frequently in human B lymphoid tumors, while N-myc gene amplification is frequent in human neuroblastomas. The observed association between aberrations in particular MYC family genes and specific subsets of malignancies might reflect, at least in part, tissue-specific differences in expression or function of a given MYC gene. Since c-Myc and N-myc share substantial functional redundancy, another factor that could influence tumor-specific gene activation would be mechanisms that target aberrations (e.g., translocations) in a given MYC gene in a particular tumor progenitor cell type. We have previously shown that mice deficient for the DNA Ligase4 (Lig4) nonhomologous DNA end-joining factor and the p53 tumor suppressor routinely develop progenitor (pro)-B cell lymphomas that harbor translocations leading to c-Myc amplification. Here, we report that a modified allele in which the c-Myc coding sequence is replaced by N-myc coding sequence (NCR allele) competes well with the wild-type c-Myc allele as a target for oncogenic translocations and amplifications in the Lig4/p53-deficient pro-B cell lymphoma model. Tumor onset, type, and cytological aberrations are similar in tumors harboring either the wild-type c-Myc gene or the NCR allele. Our results support the notion that particular features of the c-Myc locus select it as a preferential translocation/amplification target, compared to the endogenous N-myc locus, in Lig4/p53-deficient pro-B cell lymphomas.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
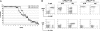
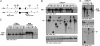

Similar articles
-
Evidence for replicative repair of DNA double-strand breaks leading to oncogenic translocation and gene amplification.J Exp Med. 2002 Aug 19;196(4):469-80. doi: 10.1084/jem.20020851. J Exp Med. 2002. PMID: 12186839 Free PMC article.
-
Oncogenic transformation in the absence of Xrcc4 targets peripheral B cells that have undergone editing and switching.J Exp Med. 2008 Dec 22;205(13):3079-90. doi: 10.1084/jem.20082271. Epub 2008 Dec 8. J Exp Med. 2008. PMID: 19064702 Free PMC article.
-
Artemis and p53 cooperate to suppress oncogenic N-myc amplification in progenitor B cells.Proc Natl Acad Sci U S A. 2004 Feb 24;101(8):2410-5. doi: 10.1073/pnas.0308757101. Proc Natl Acad Sci U S A. 2004. PMID: 14983023 Free PMC article.
-
Biology of the lymphomas: cytogenetics, molecular biology, and virology.Curr Opin Oncol. 1991 Oct;3(5):806-12. doi: 10.1097/00001622-199110000-00002. Curr Opin Oncol. 1991. PMID: 1661167 Review.
-
Constitutive activation of oncogenes by chromosomal translocations in B-cell derived tumors.AIDS Res. 1986 Dec;2 Suppl 1:S167-76. AIDS Res. 1986. PMID: 3030345 Review.
Cited by
-
Potential role of the OVOL1-OVOL2 axis and c-Myc in the progression of cutaneous squamous cell carcinoma.Mod Pathol. 2017 Jul;30(7):919-927. doi: 10.1038/modpathol.2016.169. Epub 2017 Mar 24. Mod Pathol. 2017. PMID: 28339425
-
Oncogenic Myc translocations are independent of chromosomal location and orientation of the immunoglobulin heavy chain locus.Proc Natl Acad Sci U S A. 2012 Aug 21;109(34):13728-32. doi: 10.1073/pnas.1202882109. Epub 2012 Aug 6. Proc Natl Acad Sci U S A. 2012. PMID: 22869734 Free PMC article.
-
Mechanisms of programmed DNA lesions and genomic instability in the immune system.Cell. 2013 Jan 31;152(3):417-29. doi: 10.1016/j.cell.2013.01.007. Cell. 2013. PMID: 23374339 Free PMC article. Review.
-
Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells.Cell. 2011 Sep 30;147(1):107-19. doi: 10.1016/j.cell.2011.07.049. Cell. 2011. PMID: 21962511 Free PMC article.
-
Aberrantly resolved RAG-mediated DNA breaks in Atm-deficient lymphocytes target chromosomal breakpoints in cis.Proc Natl Acad Sci U S A. 2009 Oct 27;106(43):18339-44. doi: 10.1073/pnas.0902545106. Epub 2009 Oct 9. Proc Natl Acad Sci U S A. 2009. PMID: 19820166 Free PMC article.
References
-
- Henriksson M, Luscher B. Proteins of the Myc network: essential regulators of cell growth and differentiation. Adv Cancer Res. 1996;68:109–182. - PubMed
-
- Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–699. - PubMed
-
- Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–3016. - PubMed
-
- Strieder V, Lutz W. Regulation of N-myc expression in development and disease. Cancer Lett. 2002;180:107–119. - PubMed
-
- Vita M, Henriksson M. The Myc oncoprotein as a therapeutic target for human cancer. Semin Cancer Biol. 2006;16:318–330. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

