BRG1 requirement for long-range interaction of a locus control region with a downstream promoter
- PMID: 19171905
- PMCID: PMC2650142
- DOI: 10.1073/pnas.0806420106
BRG1 requirement for long-range interaction of a locus control region with a downstream promoter
Abstract
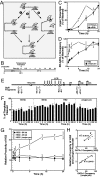
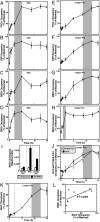
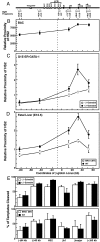
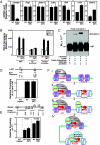
The dynamic packaging of DNA into chromatin is a fundamental step in the control of diverse nuclear processes. Whereas certain transcription factors and chromosomal components promote the formation of higher-order chromatin loops, the co-regulator machinery mediating loop assembly and disassembly is unknown. Using mice bearing a hypomorphic allele of the BRG1 chromatin remodeler, we demonstrate that the Brg1 mutation abrogated a cell type-specific loop between the beta-globin locus control region and the downstream beta major promoter, despite trans-acting factor occupancy at both sites. By contrast, distinct loops were insensitive to the Brg1 mutation. Molecular analysis with a conditional allele of GATA-1, a key regulator of hematopoiesis, in a novel cell-based system provided additional evidence that BRG1 functions early in chromatin domain activation to mediate looping. Although the paradigm in which chromatin remodelers induce nucleosome structural transitions is well established, our results demonstrating an essential role of BRG1 in the genesis of specific chromatin loops expands the repertoire of their functions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Dissecting molecular steps in chromatin domain activation during hematopoietic differentiation.Mol Cell Biol. 2007 Jun;27(12):4551-65. doi: 10.1128/MCB.00235-07. Epub 2007 Apr 16. Mol Cell Biol. 2007. PMID: 17438135 Free PMC article.
-
BRG1 directly regulates nucleosome structure and chromatin looping of the alpha globin locus to activate transcription.Nucleic Acids Res. 2009 Oct;37(18):6019-27. doi: 10.1093/nar/gkp677. Epub 2009 Aug 20. Nucleic Acids Res. 2009. PMID: 19696073 Free PMC article.
-
IUGR increases chromatin-remodeling factor Brg1 expression and binding to GR exon 1.7 promoter in newborn male rat hippocampus.Am J Physiol Regul Integr Comp Physiol. 2015 Jul 15;309(2):R119-27. doi: 10.1152/ajpregu.00495.2014. Epub 2015 May 13. Am J Physiol Regul Integr Comp Physiol. 2015. PMID: 25972460
-
The BRG1 transcriptional coregulator.Nucl Recept Signal. 2008 Feb 1;6:e004. doi: 10.1621/nrs.06004. Nucl Recept Signal. 2008. PMID: 18301784 Free PMC article. Review.
-
The BRG1 ATPase of human SWI/SNF chromatin remodeling enzymes as a driver of cancer.Epigenomics. 2017 Jun;9(6):919-931. doi: 10.2217/epi-2017-0034. Epub 2017 May 19. Epigenomics. 2017. PMID: 28521512 Free PMC article. Review.
Cited by
-
Analysis of long-range chromatin interactions using Chromosome Conformation Capture.Methods. 2012 Nov;58(3):192-203. doi: 10.1016/j.ymeth.2012.07.022. Epub 2012 Aug 15. Methods. 2012. PMID: 22903059 Free PMC article.
-
CTCF-mediated transcriptional regulation through cell type-specific chromosome organization in the β-globin locus.Nucleic Acids Res. 2012 Sep;40(16):7718-27. doi: 10.1093/nar/gks536. Epub 2012 Jun 16. Nucleic Acids Res. 2012. PMID: 22705794 Free PMC article.
-
Application of the 3C Method to Study the Developmental Genes in Drosophila Larvae.Front Genet. 2022 Jul 15;13:734208. doi: 10.3389/fgene.2022.734208. eCollection 2022. Front Genet. 2022. PMID: 35910225 Free PMC article.
-
The emerging role of chromatin remodelers in neurodevelopmental disorders: a developmental perspective.Cell Mol Life Sci. 2021 Mar;78(6):2517-2563. doi: 10.1007/s00018-020-03714-5. Epub 2020 Dec 2. Cell Mol Life Sci. 2021. PMID: 33263776 Free PMC article. Review.
-
ATP-dependent chromatin remodeling during mammalian development.Development. 2016 Aug 15;143(16):2882-97. doi: 10.1242/dev.128892. Development. 2016. PMID: 27531948 Free PMC article. Review.
References
-
- Hager GL, et al. Influence of chromatin structure on the binding of transcription factors to DNA. Cold Spring Harb Symp Quant Biol. 1993;58:63–71. - PubMed
-
- Orlando V, Strutt H, Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11:205–214. - PubMed
-
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. - PubMed
-
- Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421:448–453. - PubMed
-
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

