A general mechanism for transcription regulation by Oct1 and Oct4 in response to genotoxic and oxidative stress
- PMID: 19171782
- PMCID: PMC2648538
- DOI: 10.1101/gad.1750709
A general mechanism for transcription regulation by Oct1 and Oct4 in response to genotoxic and oxidative stress
Abstract
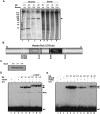
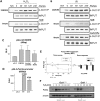
Oct1 and Oct4 are homologous transcription factors with similar DNA-binding specificities. Here we show that Oct1 is dynamically phosphorylated in vivo following exposure of cells to oxidative and genotoxic stress. We further show that stress regulates the selectivity of both proteins for specific DNA sequences. Mutation of conserved phosphorylation target DNA-binding domain residues in Oct1, and Oct4 confirms their role in regulating binding selectivity. Using chromatin immunoprecipitation, we show that association of Oct4 and Oct1 with a distinct group of in vivo targets is inducible by stress, and that Oct1 is essential for a normal post-stress transcriptional response. Finally, using an unbiased Oct1 target screen we identify a large number of genes targeted by Oct1 specifically under conditions of stress, and show that several of these inducible Oct1 targets are also inducibly bound by Oct4 in embryonic stem cells following stress exposure.
Figures


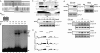

Similar articles
-
Enforcement of developmental lineage specificity by transcription factor Oct1.Elife. 2017 May 24;6:e20937. doi: 10.7554/eLife.20937. Elife. 2017. PMID: 28537559 Free PMC article.
-
Stem cells, stress, metabolism and cancer: a drama in two Octs.Trends Biochem Sci. 2009 Oct;34(10):491-9. doi: 10.1016/j.tibs.2009.06.003. Epub 2009 Sep 4. Trends Biochem Sci. 2009. PMID: 19733480 Review.
-
VIH from the mud crab is specifically expressed in the eyestalk and potentially regulated by transactivator of Sox9/Oct4/Oct1.Gen Comp Endocrinol. 2018 Jan 1;255:1-11. doi: 10.1016/j.ygcen.2017.09.018. Epub 2017 Sep 19. Gen Comp Endocrinol. 2018. PMID: 28935584
-
The Oct1 transcription factor and epithelial malignancies: Old protein learns new tricks.Biochim Biophys Acta. 2016 Jun;1859(6):792-804. doi: 10.1016/j.bbagrm.2016.02.007. Epub 2016 Feb 10. Biochim Biophys Acta. 2016. PMID: 26877236 Free PMC article. Review.
-
Phosphorylation regulates human OCT4.Proc Natl Acad Sci U S A. 2012 May 8;109(19):7162-8. doi: 10.1073/pnas.1203874109. Epub 2012 Apr 2. Proc Natl Acad Sci U S A. 2012. PMID: 22474382 Free PMC article.
Cited by
-
ChIPBase: a database for decoding the transcriptional regulation of long non-coding RNA and microRNA genes from ChIP-Seq data.Nucleic Acids Res. 2013 Jan;41(Database issue):D177-87. doi: 10.1093/nar/gks1060. Epub 2012 Nov 17. Nucleic Acids Res. 2013. PMID: 23161675 Free PMC article.
-
Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence.Genome Res. 2013 Feb;23(2):270-80. doi: 10.1101/gr.141028.112. Epub 2012 Nov 2. Genome Res. 2013. PMID: 23124521 Free PMC article.
-
Stress-induced enzyme activation primes murine embryonic stem cells to differentiate toward the first extraembryonic lineage.Stem Cells Dev. 2014 Dec 15;23(24):3049-64. doi: 10.1089/scd.2014.0157. Stem Cells Dev. 2014. PMID: 25144240 Free PMC article.
-
Development and Validation of a Rex1-RFP Potency Activity Reporter Assay That Quantifies Stress-Forced Potency Loss in Mouse Embryonic Stem Cells.Stem Cells Dev. 2016 Feb 15;25(4):320-8. doi: 10.1089/scd.2015.0169. Epub 2016 Jan 29. Stem Cells Dev. 2016. PMID: 26651054 Free PMC article.
-
Targeting Loss of Heterozygosity: A Novel Paradigm for Cancer Therapy.Pharmaceuticals (Basel). 2021 Jan 13;14(1):57. doi: 10.3390/ph14010057. Pharmaceuticals (Basel). 2021. PMID: 33450833 Free PMC article. Review.
References
-
- Almeida R., Almeida J., Shoshkes M., Mendes N., Mesquita P., Silva E., Van Seuningen I., Reis C.A., Santos-Silva F., David L. OCT-1 is over-expressed in intestinal metaplasia and intestinal gastric carcinomas and binds to, but does not transactivate, CDX2 in gastric cells. J. Pathol. 2005;207:396–401. - PubMed
-
- Chen Y., Currie R.W. Heat shock treatment suppresses angiotensin II-induced SP-1 and AP-1 and stimulates Oct-1 DNA-binding activity in heart. Inflamm. Res. 2005;54:338–343. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
