Variable locus length in the human genome leads to ascertainment bias in functional inference for non-coding elements
- PMID: 19168912
- PMCID: PMC2647827
- DOI: 10.1093/bioinformatics/btp043
Variable locus length in the human genome leads to ascertainment bias in functional inference for non-coding elements
Abstract
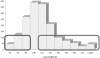
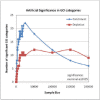
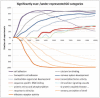
Motivation: Several functional gene annotation databases have been developed in the recent years, and are widely used to infer the biological function of gene sets, by scrutinizing the attributes that appear over- and underrepresented. However, this strategy is not directly applicable to the study of non-coding DNA, as the non-coding sequence span varies greatly among different gene loci in the human genome and longer loci have a higher likelihood of being selected purely by chance. Therefore, conclusions involving the function of non-coding elements that are drawn based on the annotation of neighboring genes are often biased. We assessed the systematic bias in several particular Gene Ontology (GO) categories using the standard hypergeometric test, by randomly sampling non-coding elements from the human genome and inferring their function based on the functional annotation of the closest genes. While no category is expected to occur significantly over- or underrepresented for a random selection of elements, categories such as 'cell adhesion', 'nervous system development' and 'transcription factor activities' appeared to be systematically overrepresented, while others such as 'olfactory receptor activity'-underrepresented.
Results: Our results suggest that functional inference for non-coding elements using gene annotation databases requires a special correction. We introduce a set of correction coefficients for the probabilities of the GO categories that accounts for the variability in the length of the non-coding DNA across different loci and effectively eliminates the ascertainment bias from the functional characterization of non-coding elements. Our approach can be easily generalized to any other gene annotation database.
Figures
Similar articles
-
[Analysis, identification and correction of some errors of model refseqs appeared in NCBI Human Gene Database by in silico cloning and experimental verification of novel human genes].Yi Chuan Xue Bao. 2004 May;31(5):431-43. Yi Chuan Xue Bao. 2004. PMID: 15478601 Chinese.
-
Integrative annotation of 21,037 human genes validated by full-length cDNA clones.PLoS Biol. 2004 Jun;2(6):e162. doi: 10.1371/journal.pbio.0020162. Epub 2004 Apr 20. PLoS Biol. 2004. PMID: 15103394 Free PMC article.
-
Properties of non-coding DNA and identification of putative cis-regulatory elements in Theileria parva.BMC Genomics. 2008 Dec 3;9:582. doi: 10.1186/1471-2164-9-582. BMC Genomics. 2008. PMID: 19055776 Free PMC article.
-
An Experimental Approach to Genome Annotation: This report is based on a colloquium sponsored by the American Academy of Microbiology held July 19-20, 2004, in Washington, DC.Washington (DC): American Society for Microbiology; 2004. Washington (DC): American Society for Microbiology; 2004. PMID: 33001599 Free Books & Documents. Review.
-
Annotation of the Drosophila melanogaster euchromatic genome: a systematic review.Genome Biol. 2002;3(12):RESEARCH0083. doi: 10.1186/gb-2002-3-12-research0083. Epub 2002 Dec 31. Genome Biol. 2002. PMID: 12537572 Free PMC article. Review.
Cited by
-
Dual transcriptional activator and repressor roles of TBX20 regulate adult cardiac structure and function.Hum Mol Genet. 2012 May 15;21(10):2194-204. doi: 10.1093/hmg/dds034. Epub 2012 Feb 10. Hum Mol Genet. 2012. PMID: 22328084 Free PMC article.
-
Improved accuracy of supervised CRM discovery with interpolated Markov models and cross-species comparison.Nucleic Acids Res. 2011 Dec;39(22):9463-72. doi: 10.1093/nar/gkr621. Epub 2011 Aug 5. Nucleic Acids Res. 2011. PMID: 21821659 Free PMC article.
-
Broad-Enrich: functional interpretation of large sets of broad genomic regions.Bioinformatics. 2014 Sep 1;30(17):i393-400. doi: 10.1093/bioinformatics/btu444. Bioinformatics. 2014. PMID: 25161225 Free PMC article.
-
Noncoding sequences near duplicated genes evolve rapidly.Genome Biol Evol. 2010;2:518-33. doi: 10.1093/gbe/evq037. Epub 2010 Jun 29. Genome Biol Evol. 2010. PMID: 20660939 Free PMC article.
-
Length bias correction for RNA-seq data in gene set analyses.Bioinformatics. 2011 Mar 1;27(5):662-9. doi: 10.1093/bioinformatics/btr005. Epub 2011 Jan 19. Bioinformatics. 2011. PMID: 21252076 Free PMC article.
References
-
- Al-Shahrour F. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. - PubMed
-
- Angst BD. The cadherin superfamily: diversity in form and function. J. Cell Sci. 2001;114:629–641. - PubMed
-
- Beissbarth T, Speed TP. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

