Highly complex neutralization determinants on a monophyletic lineage of newly transmitted subtype C HIV-1 Env clones from India
- PMID: 19167740
- PMCID: PMC2677301
- DOI: 10.1016/j.virol.2008.12.032
Highly complex neutralization determinants on a monophyletic lineage of newly transmitted subtype C HIV-1 Env clones from India
Abstract
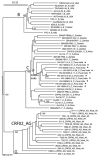
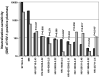
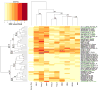
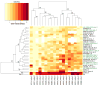
Little is known about the neutralization properties of HIV-1 in India to optimally design and test vaccines. For this reason, a functional Env clone was obtained from each of ten newly acquired, heterosexually transmitted HIV-1 infections in Pune, Maharashtra. These clones formed a phylogenetically distinct genetic lineage within subtype C. As Env-pseudotyped viruses the clones were mostly resistant to IgG1b12, 2G12 and 2F5 but all were sensitive to 4E10. When compared to a large multi-subtype panel of Env-pseudotyped viruses (subtypes B, C and CRF02_AG) in neutralization assays with a multi-subtype panel of HIV-1-positive plasma samples, the Indian Envs were remarkably complex. With the exception of the Indian Envs, results of a hierarchical clustering analysis showed a strong subtype association with the patterns of neutralization susceptibility. From these patterns we were able to identify 19 neutralization cluster-associated amino acid signatures in gp120 and 14 signatures in the ectodomain and cytoplasmic tail of gp41. We conclude that newly transmitted Indian Envs are antigenically complex in spite of close genetic similarity. Delineation of neutralization-associated amino acid signatures provides a deeper understanding of the antigenic structure of HIV-1 Env.
Figures





Similar articles
-
Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in Southern Africa.J Virol. 2006 Dec;80(23):11776-90. doi: 10.1128/JVI.01730-06. Epub 2006 Sep 13. J Virol. 2006. PMID: 16971434 Free PMC article.
-
Genetic and neutralization properties of HIV-1 env clones from subtype B/BC/AE infections in China.J Acquir Immune Defic Syndr. 2008 Apr 15;47(5):535-43. doi: 10.1097/QAI.0b013e3181663967. J Acquir Immune Defic Syndr. 2008. PMID: 18209676
-
Variations in autologous neutralization and CD4 dependence of b12 resistant HIV-1 clade C env clones obtained at different time points from antiretroviral naïve Indian patients with recent infection.Retrovirology. 2010 Sep 22;7:76. doi: 10.1186/1742-4690-7-76. Retrovirology. 2010. PMID: 20860805 Free PMC article.
-
Neutralization and infectivity characteristics of envelope glycoproteins from human immunodeficiency virus type 1 infected donors whose sera exhibit broadly cross-reactive neutralizing activity.Virology. 2006 Mar 30;347(1):36-51. doi: 10.1016/j.virol.2005.11.019. Epub 2005 Dec 27. Virology. 2006. PMID: 16378633
-
Identification of Novel Structural Determinants in MW965 Env That Regulate the Neutralization Phenotype and Conformational Masking Potential of Primary HIV-1 Isolates.J Virol. 2018 Feb 12;92(5):e01779-17. doi: 10.1128/JVI.01779-17. Print 2018 Mar 1. J Virol. 2018. PMID: 29237828 Free PMC article.
Cited by
-
Association of enhanced HIV-1 neutralization by a single Y681H substitution in gp41 with increased gp120-CD4 interaction and macrophage infectivity.PLoS One. 2012;7(5):e37157. doi: 10.1371/journal.pone.0037157. Epub 2012 May 14. PLoS One. 2012. PMID: 22606344 Free PMC article.
-
The implications of patterns in HIV diversity for neutralizing antibody induction and susceptibility.Curr Opin HIV AIDS. 2009 Sep;4(5):408-17. doi: 10.1097/COH.0b013e32832f129e. Curr Opin HIV AIDS. 2009. PMID: 20048705 Free PMC article. Review.
-
Broad and potent cross clade neutralizing antibodies with multiple specificities in the plasma of HIV-1 subtype C infected individuals.Sci Rep. 2017 Apr 24;7:46557. doi: 10.1038/srep46557. Sci Rep. 2017. PMID: 28436427 Free PMC article. Clinical Trial.
-
Rapid escape from preserved cross-reactive neutralizing humoral immunity without loss of viral fitness in HIV-1-infected progressors and long-term nonprogressors.J Virol. 2010 Apr;84(7):3576-85. doi: 10.1128/JVI.02622-09. Epub 2010 Jan 13. J Virol. 2010. PMID: 20071586 Free PMC article.
-
Tenofovir-tethered gold nanoparticles as a novel multifunctional long-acting anti-HIV therapy to overcome deficient drug delivery-: an in vivo proof of concept.J Nanobiotechnology. 2023 Jan 19;21(1):19. doi: 10.1186/s12951-022-01750-w. J Nanobiotechnology. 2023. PMID: 36658575 Free PMC article.
References
-
- Agnihotri KD, Tripathy SP, Jere AJ, Kale SM, Paranjape RS. Molecular analysis of gp41 sequences of HIV-1 type 1 subtype C from India. J Acquir Immune Defic Syndr. 2006;41:345–351. - PubMed
-
- Agnihorti K, Tripathy S, Jere A, Jadhav S, Kurle S, Paranjape R. gp120 sequences from HIV type 1 subtype C early seroconverters in India. AIDS Res Hum Retroviruses. 2004;20:889–894. - PubMed
-
- Barbato G, Bianchi E, Ingallinella P, Hurn WH, Miller MD, Ciliberto G, Cortese R, Bazzo R, Shiver JW, Pessi A. Structural analysis of the epitope of the anti-HIV antibody 2F5 sheds light into its mechanism of neutralization and HIV fusion. J Mol Biol. 2003;330:1101–1115. - PubMed
-
- Baskar PV, Ray SC, Rao R, Quinn TC, Hildreth JE, Bollinger RC. Presence in India of HIV type 1 similar to North American strains. AIDS Res Hum Retrovir. 1994;10:1039–1041. - PubMed
-
- Berlioz-Torrent C, Shacklett BL, Erdtmann L, Delamarre L, Bouchaert I, Sonigo P, Dokhelar MC, Benarous R. Interactions of the cytoplasmic domains of human and simian retroviral transmembrane proteins with components of the clathrin adaptor complexs modulate intracellular and cell surface expression of envelope glycoproteins. J Virol. 1999;73:1350–1361. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

