Differential neutralization of human immunodeficiency virus (HIV) replication in autologous CD4 T cells by HIV-specific cytotoxic T lymphocytes
- PMID: 19158248
- PMCID: PMC2655558
- DOI: 10.1128/JVI.02073-08
Differential neutralization of human immunodeficiency virus (HIV) replication in autologous CD4 T cells by HIV-specific cytotoxic T lymphocytes
Abstract
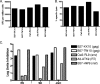
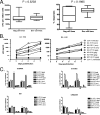
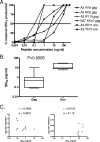
Defining the antiviral efficacy of CD8 T cells is important for immunogen design, and yet most current assays do not measure the ability of responses to neutralize infectious virus. Here we show that human immunodeficiency virus (HIV)-specific cytotoxic T-lymphocyte (CTL) clones and cell lines derived from infected persons and targeting diverse epitopes differ by over 1,000-fold in their ability to retard infectious virus replication in autologous CD4 T cells during a 7-day period in vitro, despite comparable activity as assessed by gamma interferon (IFN-gamma) enzyme-linked immunospot (ELISPOT) assay. Cell lines derived from peripheral blood mononuclear cells stimulated in vitro with peptides representing targeted Gag epitopes consistently neutralized HIV better than Env-specific lines from the same person, although ineffective inhibition of virus replication is not a universal characteristic of Env-specific responses at the clonal level. Gag-specific cell lines were of higher avidity than Env-specific lines, although avidity did not correlate with the ability of Gag- or Env-specific lines to contain HIV replication. The greatest inhibition was observed with cell lines restricted by the protective HLA alleles B*27 and B*57, but stimulation with targeted Gag epitopes resulted in greater inhibition than did stimulation with targeted Env epitopes even in non-B*27/B*57 subjects. These results assessing functional virus neutralization by HIV-specific CD8 T cells indicate that there are marked epitope- and allele-specific differences in virus neutralization by in vitro-expanded CD8 T cells, a finding not revealed by standard IFN-gamma ELISPOT assay currently in use in vaccine trials, which may be of critical importance in immunogen design and testing of candidate AIDS vaccines.
Figures

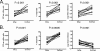

Similar articles
-
Comprehensive screening for human immunodeficiency virus type 1 subtype-specific CD8 cytotoxic T lymphocytes and definition of degenerate epitopes restricted by HLA-A0207 and -C(W)0304 alleles.J Virol. 2002 May;76(10):4971-86. doi: 10.1128/jvi.76.10.4971-4986.2002. J Virol. 2002. PMID: 11967314 Free PMC article.
-
Human immunodeficiency virus type 1-specific cytotoxic T lymphocytes release gamma interferon, tumor necrosis factor alpha (TNF-alpha), and TNF-beta when they encounter their target antigens.J Virol. 1993 May;67(5):2844-52. doi: 10.1128/JVI.67.5.2844-2852.1993. J Virol. 1993. PMID: 7682629 Free PMC article.
-
The antiviral efficacy of simian immunodeficiency virus-specific CD8+ T cells is unrelated to epitope specificity and is abrogated by viral escape.J Virol. 2007 Mar;81(6):2624-34. doi: 10.1128/JVI.01912-06. Epub 2006 Dec 27. J Virol. 2007. PMID: 17192314 Free PMC article.
-
Cytolytic T lymphocytes (CTLs) from HIV-1 subtype C-infected Indian patients recognize CTL epitopes from a conserved immunodominant region of HIV-1 Gag and Nef.J Infect Dis. 2005 Sep 1;192(5):749-59. doi: 10.1086/432547. Epub 2005 Jul 27. J Infect Dis. 2005. PMID: 16088824
-
Identification of highly conserved and broadly cross-reactive HIV type 1 cytotoxic T lymphocyte epitopes as candidate immunogens for inclusion in Mycobacterium bovis BCG-vectored HIV vaccines.AIDS Res Hum Retroviruses. 2000 Sep 20;16(14):1433-43. doi: 10.1089/08892220050140982. AIDS Res Hum Retroviruses. 2000. PMID: 11018863 Review.
Cited by
-
Characterization of CD8+ T cell differentiation following SIVΔnef vaccination by transcription factor expression profiling.PLoS Pathog. 2015 Mar 13;11(3):e1004740. doi: 10.1371/journal.ppat.1004740. eCollection 2015 Mar. PLoS Pathog. 2015. PMID: 25768938 Free PMC article.
-
Identification of a Clade-Specific HLA-C*03:02 CTL Epitope GY9 Derived from the HIV-1 p17 Matrix Protein.Int J Mol Sci. 2024 Sep 6;25(17):9683. doi: 10.3390/ijms25179683. Int J Mol Sci. 2024. PMID: 39273630 Free PMC article.
-
Gag-specific cellular immunity determines in vitro viral inhibition and in vivo virologic control following simian immunodeficiency virus challenges of vaccinated rhesus monkeys.J Virol. 2012 Sep;86(18):9583-9. doi: 10.1128/JVI.00996-12. Epub 2012 Jul 3. J Virol. 2012. PMID: 22761379 Free PMC article.
-
Early HLA-B*57-restricted CD8+ T lymphocyte responses predict HIV-1 disease progression.J Virol. 2012 Oct;86(19):10505-16. doi: 10.1128/JVI.00102-12. Epub 2012 Jul 18. J Virol. 2012. PMID: 22811521 Free PMC article.
-
Correlates of protective cellular immunity revealed by analysis of population-level immune escape pathways in HIV-1.J Virol. 2012 Dec;86(24):13202-16. doi: 10.1128/JVI.01998-12. Epub 2012 Oct 10. J Virol. 2012. PMID: 23055555 Free PMC article.
References
-
- Addo, M. M., X. G. Yu, A. Rathod, D. Cohen, R. L. Eldridge, D. Strick, M. N. Johnston, C. Corcoran, A. G. Wurcel, C. A. Fitzpatrick, M. E. Feeney, W. R. Rodriguez, N. Basgoz, R. Draenert, D. R. Stone, C. Brander, P. J. Goulder, E. S. Rosenberg, M. Altfeld, and B. D. Walker. 2003. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J. Virol. 772081-2092. - PMC - PubMed
-
- Allen, T. M., M. Altfeld, X. G. Yu, K. M. O'Sullivan, M. Lichterfeld, S. Le Gall, M. John, B. R. Mothe, P. K. Lee, E. T. Kalife, D. E. Cohen, K. A. Freedberg, D. A. Strick, M. N. Johnston, A. Sette, E. S. Rosenberg, S. A. Mallal, P. J. R. Goulder, C. Brander, and B. D. Walker. 2004. Selection, transmission, and reversion of an antigen-processing cytotoxic T-lymphocyte escape mutation in human immunodeficiency virus type 1 infection. J. Virol. 787069-7078. - PMC - PubMed
-
- Bennett, M. S., H. L. Ng, A. Ali, and O. O. Yang. 2008. Cross-clade detection of HIV-1-specific cytotoxic T lymphocytes does not reflect cross-clade antiviral activity. J. Infect. Dis. 197390-397. - PubMed
-
- Betts, M. R., D. R. Ambrozak, D. C. Douek, S. Bonhoeffer, J. M. Brenchley, J. P. Casazza, R. A. Koup, and L. J. Picker. 2001. Analysis of total human immunodeficiency virus (HIV)-specific CD4+ and CD8+ T-cell responses: relationship to viral load in untreated HIV infection. J. Virol. 7511983-11991. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

