A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis
- PMID: 19151223
- PMCID: PMC2648087
- DOI: 10.1105/tpc.108.064097
A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis
Abstract
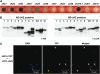
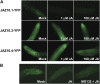
JASMONATE ZIM-domain (JAZ) proteins act as repressors of jasmonate (JA) signaling. Perception of bioactive JAs by the F-box protein CORONATINE INSENSITIVE1 (COI1) causes degradation of JAZs via the ubiquitin-proteasome pathway, which in turn activates the expression of genes involved in plant growth, development, and defense. JAZ proteins contain two highly conserved sequence regions: the Jas domain that interacts with COI1 to destabilize the repressor and the ZIM domain of unknown function. Here, we show that the conserved TIFY motif (TIFF/YXG) within the ZIM domain mediates homo- and heteromeric interactions between most Arabidopsis thaliana JAZs. We have also identified an alternatively spliced form (JAZ10.4) of JAZ10 that lacks the Jas domain and, as a consequence, is highly resistant to JA-induced degradation. Strong JA-insensitive phenotypes conferred by overexpression of JAZ10.4 were suppressed by mutations in the TIFY motif that block JAZ10.4-JAZ interactions. We conclude that JAZ10.4 functions to attenuate signal output in the presence of JA and further suggest that the dominant-negative action of this splice variant involves protein-protein interaction through the ZIM/TIFY domain. The ability of JAZ10.4 to interact with MYC2 is consistent with a model in which a JAZ10.4-containing protein complex directly represses the activity of transcription factors that promote expression of JA response genes.
Figures






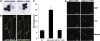
Similar articles
-
Negative feedback control of jasmonate signaling by an alternative splice variant of JAZ10.Plant Physiol. 2013 Jun;162(2):1006-17. doi: 10.1104/pp.113.218164. Epub 2013 Apr 30. Plant Physiol. 2013. PMID: 23632853 Free PMC article.
-
A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein.Plant J. 2008 Sep;55(6):979-88. doi: 10.1111/j.1365-313X.2008.03566.x. Epub 2008 Jun 10. Plant J. 2008. PMID: 18547396 Free PMC article.
-
Structural insights into alternative splicing-mediated desensitization of jasmonate signaling.Proc Natl Acad Sci U S A. 2017 Feb 14;114(7):1720-1725. doi: 10.1073/pnas.1616938114. Epub 2017 Jan 30. Proc Natl Acad Sci U S A. 2017. PMID: 28137867 Free PMC article.
-
The JAZ proteins: a crucial interface in the jasmonate signaling cascade.Plant Cell. 2011 Sep;23(9):3089-100. doi: 10.1105/tpc.111.089300. Epub 2011 Sep 30. Plant Cell. 2011. PMID: 21963667 Free PMC article. Review.
-
JAZ repressors set the rhythm in jasmonate signaling.Curr Opin Plant Biol. 2008 Oct;11(5):486-94. doi: 10.1016/j.pbi.2008.06.003. Epub 2008 Jul 22. Curr Opin Plant Biol. 2008. PMID: 18653378 Review.
Cited by
-
NaJAZh regulates a subset of defense responses against herbivores and spontaneous leaf necrosis in Nicotiana attenuata plants.Plant Physiol. 2012 Jun;159(2):769-88. doi: 10.1104/pp.112.193771. Epub 2012 Apr 9. Plant Physiol. 2012. PMID: 22496510 Free PMC article.
-
A structure-redesigned intrinsically disordered peptide that selectively inhibits a plant transcription factor in jasmonate signaling.PNAS Nexus. 2024 Jul 26;3(8):pgae312. doi: 10.1093/pnasnexus/pgae312. eCollection 2024 Aug. PNAS Nexus. 2024. PMID: 39139264 Free PMC article.
-
Transcriptome Analysis Reveals Changes in Whole Gene Expression, Biological Process, and Molecular Functions Induced by Nickel in Jack Pine (Pinus banksiana).Plants (Basel). 2023 Aug 7;12(15):2889. doi: 10.3390/plants12152889. Plants (Basel). 2023. PMID: 37571042 Free PMC article.
-
JA modulates phytochrome a signaling via repressing FHY3 activity by JAZ proteins.Plant Signal Behav. 2020 Mar 3;15(3):1726636. doi: 10.1080/15592324.2020.1726636. Epub 2020 Feb 11. Plant Signal Behav. 2020. PMID: 32043408 Free PMC article.
-
Responses of differential metabolites and pathways to high temperature in cucumber anther.Front Plant Sci. 2023 Apr 14;14:1131735. doi: 10.3389/fpls.2023.1131735. eCollection 2023. Front Plant Sci. 2023. PMID: 37123826 Free PMC article.
References
-
- Balbi, V., and Devoto, A. (2008). Jasmonate signalling network in Arabidopsis thaliana: Crucial regulatory nodes and new physiological scenarios. New Phytol. 177 301–318. - PubMed
-
- Barbazuk, W.B., Fu, Y., and McGinnis, K.M. (2008). Genome-wide analyses of alternative splicing in plants: Opportunities and challenges. Genome Res. 18 1381–1392. - PubMed
-
- Bove, J., Kim, C.Y., Gibson, C.A., and Assmann, S.M. (2008). Characterization of wound-responsive RNA-binding proteins and their splice variants in Arabidopsis. Plant Mol. Biol. 67 71–88. - PubMed
-
- Bracha-Drori, K., Shichrur, K., Katz, A., Oliva, M., Angelovici, R., Yalovsky, S., and Ohad, N. (2004). Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J. 40 419–427. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

