Yeast genetic analysis reveals the involvement of chromatin reassembly factors in repressing HIV-1 basal transcription
- PMID: 19148280
- PMCID: PMC2613532
- DOI: 10.1371/journal.pgen.1000339
Yeast genetic analysis reveals the involvement of chromatin reassembly factors in repressing HIV-1 basal transcription
Erratum in
- PLoS Genet. 2009 Jan;5(1). doi: 10.1371/annotation/45587774-5d7c-4023-97b0-96833be7ad9a
Abstract
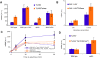
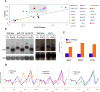
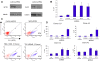
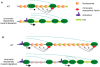
Rebound of HIV viremia after interruption of anti-retroviral therapy is due to the small population of CD4+ T cells that remain latently infected. HIV-1 transcription is the main process controlling post-integration latency. Regulation of HIV-1 transcription takes place at both initiation and elongation levels. Pausing of RNA polymerase II at the 5' end of HIV-1 transcribed region (5'HIV-TR), which is immediately downstream of the transcription start site, plays an important role in the regulation of viral expression. The activation of HIV-1 transcription correlates with the rearrangement of a positioned nucleosome located at this region. These two facts suggest that the 5'HIV-TR contributes to inhibit basal transcription of those HIV-1 proviruses that remain latently inactive. However, little is known about the cell elements mediating the repressive role of the 5'HIV-TR. We performed a genetic analysis of this phenomenon in Saccharomyces cerevisiae after reconstructing a minimal HIV-1 transcriptional system in this yeast. Unexpectedly, we found that the critical role played by the 5'HIV-TR in maintaining low levels of basal transcription in yeast is mediated by FACT, Spt6, and Chd1, proteins so far associated with chromatin assembly and disassembly during ongoing transcription. We confirmed that this group of factors plays a role in HIV-1 postintegration latency in human cells by depleting the corresponding human orthologs with shRNAs, both in HIV latently infected cell populations and in particular single-integration clones, including a latent clone with a provirus integrated in a highly transcribed gene. Our results indicate that chromatin reassembly factors participate in the establishment of the equilibrium between activation and repression of HIV-1 when it integrates into the human genome, and they open the possibility of considering these factors as therapeutic targets of HIV-1 latency.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Negative elongation factor (NELF) coordinates RNA polymerase II pausing, premature termination, and chromatin remodeling to regulate HIV transcription.J Biol Chem. 2013 Sep 6;288(36):25995-26003. doi: 10.1074/jbc.M113.496489. Epub 2013 Jul 24. J Biol Chem. 2013. PMID: 23884411 Free PMC article.
-
Chromatin reassembly factors are involved in transcriptional interference promoting HIV latency.J Virol. 2011 Apr;85(7):3187-202. doi: 10.1128/JVI.01920-10. Epub 2011 Jan 26. J Virol. 2011. PMID: 21270164 Free PMC article.
-
Negative elongation factor is required for the maintenance of proviral latency but does not induce promoter-proximal pausing of RNA polymerase II on the HIV long terminal repeat.Mol Cell Biol. 2014 Jun;34(11):1911-28. doi: 10.1128/MCB.01013-13. Epub 2014 Mar 17. Mol Cell Biol. 2014. PMID: 24636995 Free PMC article.
-
Chromatin-associated regulation of HIV-1 transcription: implications for the development of therapeutic strategies.Subcell Biochem. 2007;41:371-96. Subcell Biochem. 2007. PMID: 17484137 Review.
-
Factors controlling chromatin organization and nucleosome positioning for establishment and maintenance of HIV latency.Curr HIV Res. 2008 Jun;6(4):286-95. doi: 10.2174/157016208785132563. Curr HIV Res. 2008. PMID: 18691027 Review.
Cited by
-
The impact of the HIRA histone chaperone upon global nucleosome architecture.Cell Cycle. 2015;14(1):123-34. doi: 10.4161/15384101.2014.967123. Cell Cycle. 2015. PMID: 25602522 Free PMC article.
-
Crystal structures of the S. cerevisiae Spt6 core and C-terminal tandem SH2 domain.J Mol Biol. 2011 May 13;408(4):697-713. doi: 10.1016/j.jmb.2011.03.002. Epub 2011 Mar 17. J Mol Biol. 2011. PMID: 21419780 Free PMC article.
-
HIV interactions with monocytes and dendritic cells: viral latency and reservoirs.Retrovirology. 2009 Jun 1;6:51. doi: 10.1186/1742-4690-6-51. Retrovirology. 2009. PMID: 19486514 Free PMC article. Review.
-
CHD1 and CHD2 are positive regulators of HIV-1 gene expression.Virol J. 2014 Oct 8;11:180. doi: 10.1186/1743-422X-11-180. Virol J. 2014. PMID: 25297984 Free PMC article.
-
The ribosome assembly gene network is controlled by the feedback regulation of transcription elongation.Nucleic Acids Res. 2017 Sep 19;45(16):9302-9318. doi: 10.1093/nar/gkx529. Nucleic Acids Res. 2017. PMID: 28637236 Free PMC article.
References
-
- Palangat M, Meier TI, Keene RG, Landick R. Transcriptional pausing at +62 of the HIV-1 nascent RNA modulates formation of the TAR RNA structure. Mol Cell. 1998;1:1033–1042. - PubMed
-
- Kao SY, Calman AF, Luciw PA, Peterlin BM. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987;330:489–493. - PubMed
-
- Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. - PubMed
-
- Benkirane M, Chun RF, Xiao H, Ogryzko VV, Howard BH, et al. Activation of integrated provirus requires histone acetyltransferase. p300 and P/CAF are coactivators for HIV-1 Tat. J Biol Chem. 1998;273:24898–24905. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

