Quality control of a transcriptional regulator by SUMO-targeted degradation
- PMID: 19139279
- PMCID: PMC2655623
- DOI: 10.1128/MCB.01470-08
Quality control of a transcriptional regulator by SUMO-targeted degradation
Abstract
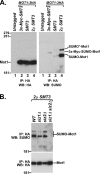
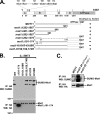
Slx5 and Slx8 are heterodimeric RING domain-containing proteins that possess SUMO-targeted ubiquitin ligase (STUbL) activity in vitro. Slx5-Slx8 and its orthologs are proposed to target SUMO conjugates for ubiquitin-mediated proteolysis, but the only in vivo substrate identified to date is mammalian PML, and the physiological importance of SUMO-targeted ubiquitylation remains largely unknown. We previously identified mutations in SLX5 and SLX8 by selecting for suppressors of a temperature-sensitive allele of MOT1, which encodes a regulator of TATA-binding protein. Here, we demonstrate that Mot1 is SUMOylated in vivo and that disrupting the Slx5-Slx8 pathway by mutation of the target lysines in Mot1, by deletion of SLX5 or the ubiquitin E2 UBC4, or by inhibition of the proteosome suppresses mot1-301 mutant phenotypes and increases the stability of the Mot1-301 protein. The Mot1-301 mutant protein is targeted for proteolysis by SUMOylation to a much greater extent than wild-type Mot1, suggesting a quality control mechanism. In support of this idea, growth of Saccharomyces cerevisiae in the presence of the arginine analog canavanine results in increased SUMOylation and Slx5-Slx8-mediated degradation of wild-type Mot1. These results therefore demonstrate that Mot1 is an in vivo STUbL target in yeast and suggest a role for SUMO-targeted degradation in protein quality control.
Figures

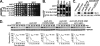

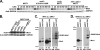

Similar articles
-
Genetic analysis connects SLX5 and SLX8 to the SUMO pathway in Saccharomyces cerevisiae.Genetics. 2006 Mar;172(3):1499-509. doi: 10.1534/genetics.105.052811. Epub 2005 Dec 30. Genetics. 2006. PMID: 16387868 Free PMC article.
-
Activation of the Slx5-Slx8 ubiquitin ligase by poly-small ubiquitin-like modifier conjugates.J Biol Chem. 2008 Jul 18;283(29):19912-21. doi: 10.1074/jbc.M802690200. Epub 2008 May 22. J Biol Chem. 2008. PMID: 18499666 Free PMC article.
-
SUMO-independent in vivo activity of a SUMO-targeted ubiquitin ligase toward a short-lived transcription factor.Genes Dev. 2010 May;24(9):893-903. doi: 10.1101/gad.1906510. Epub 2010 Apr 13. Genes Dev. 2010. PMID: 20388728 Free PMC article.
-
Nuclear organization in genome stability: SUMO connections.Cell Res. 2011 Mar;21(3):474-85. doi: 10.1038/cr.2011.31. Epub 2011 Feb 15. Cell Res. 2011. PMID: 21321608 Free PMC article. Review.
-
SUMO-targeted ubiquitin ligases.Biochim Biophys Acta. 2014 Jan;1843(1):75-85. doi: 10.1016/j.bbamcr.2013.08.022. Epub 2013 Sep 7. Biochim Biophys Acta. 2014. PMID: 24018209 Review.
Cited by
-
STUbLs in chromatin and genome stability.Biopolymers. 2013 Feb;99(2):146-54. doi: 10.1002/bip.22125. Biopolymers. 2013. PMID: 23175389 Free PMC article. Review.
-
Degradation of the Saccharomyces cerevisiae mating-type regulator alpha1: genetic dissection of cis-determinants and trans-acting pathways.Genetics. 2010 Jun;185(2):497-511. doi: 10.1534/genetics.110.115907. Epub 2010 Mar 29. Genetics. 2010. PMID: 20351217 Free PMC article.
-
Genetic evidence that polysumoylation bypasses the need for a SUMO-targeted Ub ligase.Genetics. 2011 Jan;187(1):73-87. doi: 10.1534/genetics.110.124347. Epub 2010 Nov 8. Genetics. 2011. PMID: 21059884 Free PMC article.
-
Physical and Genetic Interactions Between Uls1 and the Slx5-Slx8 SUMO-Targeted Ubiquitin Ligase.G3 (Bethesda). 2013 Apr 9;3(4):771-780. doi: 10.1534/g3.113.005827. G3 (Bethesda). 2013. PMID: 23550137 Free PMC article.
-
Role of RNF4 in the ubiquitination of Rta of Epstein-Barr virus.J Biol Chem. 2013 May 3;288(18):12866-79. doi: 10.1074/jbc.M112.413393. Epub 2013 Mar 15. J Biol Chem. 2013. PMID: 23504328 Free PMC article.
References
-
- Adamkewicz, J. I., K. E. Hansen, W. A. Prud'homme, J. L. Davis, and J. Thorner. 2001. High affinity interaction of yeast transcriptional regulator, Mot1, with TATA box-binding protein (TBP). J. Biol. Chem. 27611883-11894. - PubMed
-
- Baba, D., N. Maita, J. G. Jee, Y. Uchimura, H. Saitoh, K. Sugasawa, F. Hanaoka, H. Tochio, H. Hiroaki, and M. Shirakawa. 2005. Crystal structure of thymine DNA glycosylase conjugated to SUMO-1. Nature 435979-982. - PubMed
-
- Cardone, L., J. Hirayama, F. Giordano, T. Tamaru, J. J. Palvimo, and P. Sassone-Corsi. 2005. Circadian clock control by SUMOylation of BMAL1. Science 3091390-1394. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
