A methodology for the combined in situ analyses of the precursor and mature forms of microRNAs and correlation with their putative targets
- PMID: 19131963
- PMCID: PMC2709217
- DOI: 10.1038/nprot.2008.215
A methodology for the combined in situ analyses of the precursor and mature forms of microRNAs and correlation with their putative targets
Abstract
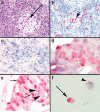
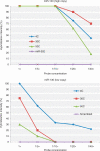
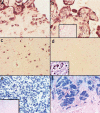
There are relatively few protocols described for the in situ detection of microRNA (miRNA) and they often use cryostat sections, signal amplification and hybridization or washes of 50-60 degrees C. This protocol describes in situ miRNA detection that can be done in paraffin-embedded, formalin-fixed tissue. Detection of the miRNA precursors can be done by RT in situ PCR, which can theoretically detect one copy per cell. The key variable for the RT in situ PCR protocol is optimal protease digestion, which is then followed by overnight DNase digestion and target specific incorporation of the reported nucleotide into the amplified cDNA. Detection of mature miRNAs is achieved by in situ hybridization with locked nucleic acid probes. This part of the protocol involves a brief protease digestion, followed by an overnight hybridization, short low stringency wash and detection of the labeled probe. The key variables for this method include probe concentration and stringency conditions. Each miRNA in situ method takes 1 d. The final step of the protocol involves colabeling by immunohistochemistry for the putative target of the miRNA, which is done after the in situ hybridization step and takes a few hours.
Figures
Similar articles
-
In situ detection of mature microRNAs by labeled extension on ultramer templates.Biotechniques. 2009 Feb;46(2):115-26. doi: 10.2144/000113068. Biotechniques. 2009. PMID: 19317656 Free PMC article.
-
Co-labeling using in situ PCR: a review.J Histochem Cytochem. 2001 Nov;49(11):1329-39. doi: 10.1177/002215540104901101. J Histochem Cytochem. 2001. PMID: 11668186 Review.
-
The foundations of successful RT in situ PCR.Front Biosci. 1996 Nov 1;1:c4-15. doi: 10.2741/a110. Front Biosci. 1996. PMID: 9159200 Review.
-
In situ detection of precursor and mature microRNAs in paraffin embedded, formalin fixed tissues and cell preparations.Methods. 2008 Jan;44(1):39-46. doi: 10.1016/j.ymeth.2007.10.008. Methods. 2008. PMID: 18158131 Review.
-
In situ detection of microRNAs in paraffin embedded, formalin fixed tissues and the co-localization of their putative targets.Methods. 2010 Dec;52(4):307-15. doi: 10.1016/j.ymeth.2010.08.009. Epub 2010 Aug 17. Methods. 2010. PMID: 20723602 Review.
Cited by
-
Small RNA Detection by in Situ Hybridization Methods.Int J Mol Sci. 2015 Jun 10;16(6):13259-86. doi: 10.3390/ijms160613259. Int J Mol Sci. 2015. PMID: 26068454 Free PMC article. Review.
-
Defining larger roles for "tiny" RNA molecules: role of miRNAs in neurodegeneration research.J Neuroimmune Pharmacol. 2010 Mar;5(1):63-9. doi: 10.1007/s11481-009-9172-4. Epub 2009 Sep 16. J Neuroimmune Pharmacol. 2010. PMID: 19757077 Free PMC article. Review.
-
In Situ Hybridization for Detecting Mature MicroRNAs In Vivo at Single-Cell Resolution.Curr Protoc Mol Biol. 2019 Jun;127(1):e93. doi: 10.1002/cpmb.93. Curr Protoc Mol Biol. 2019. PMID: 31237425 Free PMC article.
-
Prognostic value of the MicroRNA regulators Dicer and Drosha in non-small-cell lung cancer: co-expression of Drosha and miR-126 predicts poor survival.BMC Clin Pathol. 2014 Dec 11;14(1):45. doi: 10.1186/1472-6890-14-45. eCollection 2014. BMC Clin Pathol. 2014. PMID: 25525410 Free PMC article.
-
Clinical application of microRNA in gastric cancer in Eastern Asian area.World J Gastroenterol. 2013 Apr 7;19(13):2019-27. doi: 10.3748/wjg.v19.i13.2019. World J Gastroenterol. 2013. PMID: 23599620 Free PMC article. Review.
References
-
- Nuovo GJ. In situ detection of precursor and mature microRNAs in paraffin embedded, formalin fixed tissues and cell preparations. Methods. 2008;44:39–46. - PubMed
-
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. - PubMed
-
- Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. MicroRNA expression and function in cancer. Trends Mol. Med. 2006;12:580–587. - PubMed
-
- Yanaihara N, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

