The loss of the snoRNP chaperone Nopp140 from Cajal bodies of patient fibroblasts correlates with the severity of spinal muscular atrophy
- PMID: 19129172
- PMCID: PMC2655770
- DOI: 10.1093/hmg/ddp009
The loss of the snoRNP chaperone Nopp140 from Cajal bodies of patient fibroblasts correlates with the severity of spinal muscular atrophy
Abstract
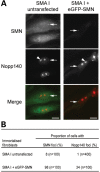
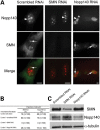
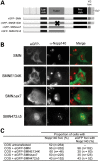
Spinal muscular atrophy (SMA) is a common autosomal recessive neurodegenerative disease caused by reduced survival motor neuron (SMN) levels. The assembly machinery containing SMN is implicated in the biogenesis of the spliceosomal small nuclear ribonucleoproteins (snRNPs). SMN is present in both the cytoplasm and nucleus, where it transiently accumulates in subnuclear domains named Cajal bodies (CBs) and functions in the maturation of snRNPs and small nucleolar (sno)RNPs. The impact of lowering SMN levels on the composition of CBs in SMA cells is still not completely understood. Here, we analyse the CB composition in immortalized and primary fibroblasts from SMA patients. We show that the U snRNA export factors PHAX and chromosome region maintenance 1 and the box C/D snoRNP core protein fibrillarin concentrate in CBs from SMA cells, whereas the box H/ACA core proteins GAR1 and NAP57/dyskerin show reduced CB localization. Remarkably, the functional deficiency in SMA cells is associated with decreased localization of the snoRNP chaperone Nopp140 in CBs that correlates with disease severity. Indeed, RNA interference knockdown experiments in control fibroblasts demonstrate that SMN is required for accumulation of Nopp140 in CBs. Conversely, overexpression of SMN in SMA cells restores the CB localization of Nopp140, whereas SMN mutants found in SMA patients are defective in promoting the association of Nopp140 with CBs. Taken together, we demonstrate that only a subset of CB functions (as indicated by the association of representative factors) are impaired in SMA cells and, importantly, we identify the decrease of Nopp140 localization in CBs as a phenotypic marker for SMA.
Figures

Similar articles
-
Distinct domains of the spinal muscular atrophy protein SMN are required for targeting to Cajal bodies in mammalian cells.J Cell Sci. 2006 Feb 15;119(Pt 4):680-92. doi: 10.1242/jcs.02782. Epub 2006 Jan 31. J Cell Sci. 2006. PMID: 16449324
-
Ongoing U snRNP biogenesis is required for the integrity of Cajal bodies.Mol Biol Cell. 2006 Jul;17(7):3221-31. doi: 10.1091/mbc.e06-03-0247. Epub 2006 May 10. Mol Biol Cell. 2006. PMID: 16687569 Free PMC article.
-
A role for protein phosphatase PP1γ in SMN complex formation and subnuclear localization to Cajal bodies.J Cell Sci. 2012 Jun 15;125(Pt 12):2862-74. doi: 10.1242/jcs.096255. Epub 2012 Mar 27. J Cell Sci. 2012. PMID: 22454514
-
RNP Assembly Defects in Spinal Muscular Atrophy.Adv Neurobiol. 2018;20:143-171. doi: 10.1007/978-3-319-89689-2_6. Adv Neurobiol. 2018. PMID: 29916019 Review.
-
SMN - A chaperone for nuclear RNP social occasions?RNA Biol. 2017 Jun 3;14(6):701-711. doi: 10.1080/15476286.2016.1236168. Epub 2016 Sep 20. RNA Biol. 2017. PMID: 27648855 Free PMC article. Review.
Cited by
-
New prospects for targeting telomerase beyond the telomere.Nat Rev Cancer. 2016 Aug;16(8):508-24. doi: 10.1038/nrc.2016.55. Epub 2016 Jun 24. Nat Rev Cancer. 2016. PMID: 27339602 Review.
-
Cajal body surveillance of U snRNA export complex assembly.J Cell Biol. 2010 Aug 23;190(4):603-12. doi: 10.1083/jcb.201004109. J Cell Biol. 2010. PMID: 20733056 Free PMC article.
-
The Small-Molecule Flunarizine in Spinal Muscular Atrophy Patient Fibroblasts Impacts on the Gemin Components of the SMN Complex and TDP43, an RNA-Binding Protein Relevant to Motor Neuron Diseases.Front Mol Biosci. 2020 Apr 17;7:55. doi: 10.3389/fmolb.2020.00055. eCollection 2020. Front Mol Biosci. 2020. PMID: 32363199 Free PMC article.
-
The role of nuclear bodies in gene expression and disease.Biology (Basel). 2013 Jul 9;2(3):976-1033. doi: 10.3390/biology2030976. Biology (Basel). 2013. PMID: 24040563 Free PMC article.
-
The SMN-ribosome interplay: a new opportunity for Spinal Muscular Atrophy therapies.Biochem Soc Trans. 2024 Feb 28;52(1):465-479. doi: 10.1042/BST20231116. Biochem Soc Trans. 2024. PMID: 38391004 Free PMC article. Review.
References
-
- Lefebvre S., Bürglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. - PubMed
-
- Lefebvre S., Burlet P., Liu Q., Bertrandy S., Clermont O., Munnich A., Dreyfuss G., Melki J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet. 1997;16:265–269. - PubMed
-
- Coovert D.D., Le T.T., McAndrew P.E., Strasswimmer J., Crawford T.O., Mendell J.R., Coulson S.E., Androphy E.J., Prior T.W., Burghes A.H. The survival motor neuron protein in spinal muscular atrophy. Hum. Mol. Genet. 1997;6:1205–1214. - PubMed
-
- Monani U.R. Spinal muscular atrophy: a deficiency in a ubiquitous protein; a motor neuron-specific disease (Review) Neuron. 2005;48:885–896. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

