Characterization of the Caenorhabditis elegans Islet LIM-homeodomain ortholog, lim-7
- PMID: 19116151
- PMCID: PMC2719984
- DOI: 10.1016/j.febslet.2008.12.046
Characterization of the Caenorhabditis elegans Islet LIM-homeodomain ortholog, lim-7
Abstract
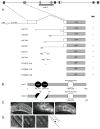
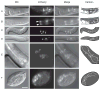
lim-7 is one of seven Caenorhabditis elegans LIM-homeodomain (LIM-HD)-encoding genes and the sole Islet ortholog. LIM-HD transcription factors, including Islets, function in neuronal and non-neuronal development across diverse phyla. Our results show that a lim-7 deletion allele causes early larval lethality with terminal phenotypes including uncoordination, detached pharynx, constipation and morphological defects. A lim-7(+) transgene rescues lethality but not adult sterility. A lim-7(+) reporter in the full genomic context is expressed in all gonadal sheath cells, URA neurons, and additional cells in the pharyngeal region. Finally, we identify a 45-bp regulatory element in the first intron that is necessary and sufficient for lim-7 gonadal sheath expression.
Figures



Similar articles
-
Neuron cell type-specific SNAP-25 expression driven by multiple regulatory elements in the nematode Caenorhabditis elegans.J Mol Biol. 2003 Oct 17;333(2):237-47. doi: 10.1016/j.jmb.2003.08.055. J Mol Biol. 2003. PMID: 14529613
-
Intron-specific patterns of divergence of lin-11 regulatory function in the C. elegans nervous system.Dev Biol. 2017 Apr 1;424(1):90-103. doi: 10.1016/j.ydbio.2017.02.005. Epub 2017 Feb 17. Dev Biol. 2017. PMID: 28215941
-
The Caenorhabditis elegans lim-6 LIM homeobox gene regulates neurite outgrowth and function of particular GABAergic neurons.Development. 1999 Apr;126(7):1547-62. doi: 10.1242/dev.126.7.1547. Development. 1999. PMID: 10068647
-
Securing Neuronal Cell Fate in C. elegans.Curr Top Dev Biol. 2016;116:167-80. doi: 10.1016/bs.ctdb.2015.11.011. Epub 2016 Jan 23. Curr Top Dev Biol. 2016. PMID: 26970619 Review.
-
Brn3/POU-IV-type POU homeobox genes-Paradigmatic regulators of neuronal identity across phylogeny.Wiley Interdiscip Rev Dev Biol. 2020 Jul;9(4):e374. doi: 10.1002/wdev.374. Epub 2020 Feb 3. Wiley Interdiscip Rev Dev Biol. 2020. PMID: 32012462 Review.
Cited by
-
GPI-anchor synthesis is indispensable for the germline development of the nematode Caenorhabditis elegans.Mol Biol Cell. 2012 Mar;23(6):982-95. doi: 10.1091/mbc.E10-10-0855. Epub 2012 Feb 1. Mol Biol Cell. 2012. PMID: 22298425 Free PMC article.
-
A lysine-rich motif in the phosphatidylserine receptor PSR-1 mediates recognition and removal of apoptotic cells.Nat Commun. 2015 Jan 7;6:5717. doi: 10.1038/ncomms6717. Nat Commun. 2015. PMID: 25564762 Free PMC article.
-
ERK phosphorylates chromosomal axis component HORMA domain protein HTP-1 to regulate oocyte numbers.Sci Adv. 2020 Oct 30;6(44):eabc5580. doi: 10.1126/sciadv.abc5580. Print 2020 Oct. Sci Adv. 2020. PMID: 33127680 Free PMC article.
-
Cell Non-autonomous Function of daf-18/PTEN in the Somatic Gonad Coordinates Somatic Gonad and Germline Development in C. elegans Dauer Larvae.Curr Biol. 2019 Mar 18;29(6):1064-1072.e8. doi: 10.1016/j.cub.2019.01.076. Epub 2019 Feb 28. Curr Biol. 2019. PMID: 30827916 Free PMC article.
-
Dauer juvenile recovery transcriptome of two contrasting EMS mutants of the entomopathogenic nematode Heterorhabditis bacteriophora.World J Microbiol Biotechnol. 2024 Mar 7;40(4):128. doi: 10.1007/s11274-024-03902-6. World J Microbiol Biotechnol. 2024. PMID: 38451353
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

