Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex
- PMID: 19109893
- PMCID: PMC2676164
- DOI: 10.1016/j.cell.2008.10.045
Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex
Abstract
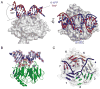
Ultraviolet (UV) light-induced pyrimidine photodimers are repaired by the nucleotide excision repair pathway. Photolesions have biophysical parameters closely resembling undamaged DNA, impeding discovery through damage surveillance proteins. The DDB1-DDB2 complex serves in the initial detection of UV lesions in vivo. Here we present the structures of the DDB1-DDB2 complex alone and bound to DNA containing either a 6-4 pyrimidine-pyrimidone photodimer (6-4PP) lesion or an abasic site. The structure shows that the lesion is held exclusively by the WD40 domain of DDB2. A DDB2 hairpin inserts into the minor groove, extrudes the photodimer into a binding pocket, and kinks the duplex by approximately 40 degrees. The tightly localized probing of the photolesions, combined with proofreading in the photodimer pocket, enables DDB2 to detect lesions refractory to detection by other damage surveillance proteins. The structure provides insights into damage recognition in chromatin and suggests a mechanism by which the DDB1-associated CUL4 ubiquitin ligase targets proteins surrounding the site of damage.
Figures
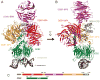
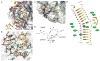
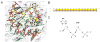
Similar articles
-
DDB1-DDB2 (xeroderma pigmentosum group E) protein complex recognizes a cyclobutane pyrimidine dimer, mismatches, apurinic/apyrimidinic sites, and compound lesions in DNA.J Biol Chem. 2005 Dec 2;280(48):39982-9. doi: 10.1074/jbc.M507854200. Epub 2005 Aug 24. J Biol Chem. 2005. PMID: 16223728
-
DNA damage binding protein component DDB1 participates in nucleotide excision repair through DDB2 DNA-binding and cullin 4A ubiquitin ligase activity.Cancer Res. 2006 Sep 1;66(17):8590-7. doi: 10.1158/0008-5472.CAN-06-1115. Cancer Res. 2006. PMID: 16951172
-
The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites.Proc Natl Acad Sci U S A. 2006 Feb 21;103(8):2588-93. doi: 10.1073/pnas.0511160103. Epub 2006 Feb 10. Proc Natl Acad Sci U S A. 2006. PMID: 16473935 Free PMC article.
-
Xeroderma pigmentosum complementation group E and UV-damaged DNA-binding protein.DNA Repair (Amst). 2002 Aug 6;1(8):601-16. doi: 10.1016/s1568-7864(02)00052-6. DNA Repair (Amst). 2002. PMID: 12509284 Free PMC article. Review.
-
Detecting UV-lesions in the genome: The modular CRL4 ubiquitin ligase does it best!FEBS Lett. 2011 Sep 16;585(18):2818-25. doi: 10.1016/j.febslet.2011.04.064. Epub 2011 May 6. FEBS Lett. 2011. PMID: 21550341 Review.
Cited by
-
Photodynamic viral inactivation: Recent advances and potential applications.Appl Phys Rev. 2021 Jun;8(2):021315. doi: 10.1063/5.0044713. Appl Phys Rev. 2021. PMID: 34084253 Free PMC article. Review.
-
PCalign: a method to quantify physicochemical similarity of protein-protein interfaces.BMC Bioinformatics. 2015 Feb 1;16:33. doi: 10.1186/s12859-015-0471-x. BMC Bioinformatics. 2015. PMID: 25638036 Free PMC article.
-
CRL4-DCAF1 Ubiquitin Ligase Dependent Functions of HIV Viral Protein R and Viral Protein X.Viruses. 2024 Aug 17;16(8):1313. doi: 10.3390/v16081313. Viruses. 2024. PMID: 39205287 Free PMC article. Review.
-
Mammalian transcription-coupled excision repair.Cold Spring Harb Perspect Biol. 2013 Aug 1;5(8):a012625. doi: 10.1101/cshperspect.a012625. Cold Spring Harb Perspect Biol. 2013. PMID: 23906714 Free PMC article. Review.
-
Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity.Nat Rev Genet. 2009 Nov;10(11):756-68. doi: 10.1038/nrg2663. Epub 2009 Oct 7. Nat Rev Genet. 2009. PMID: 19809470 Review.
References
-
- Aboussekhra A, Biggerstaff M, Shivji MK, Vilpo JA, Moncollin V, Podust VN, Protic M, Hubscher U, Egly JM, Wood RD. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 1995;80:859–868. - PubMed
-
- Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 2006;443:590–593. - PubMed
-
- Batty D, Rapic'-Otrin V, Levine AS, Wood RD. Stable binding of human XPC complex to irradiated DNA confers strong discrimination for damaged sites. J Mol Biol. 2000;300:275–290. - PubMed
-
- Cleaver JE. Cancer in xeroderma pigmentosum and related disorders of DNA repair. Nat Rev Cancer. 2005;5:564–573. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

