Deficient SUMO attachment to Flp recombinase leads to homologous recombination-dependent hyperamplification of the yeast 2 microm circle plasmid
- PMID: 19109426
- PMCID: PMC2642740
- DOI: 10.1091/mbc.e08-06-0659
Deficient SUMO attachment to Flp recombinase leads to homologous recombination-dependent hyperamplification of the yeast 2 microm circle plasmid
Abstract
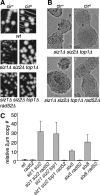
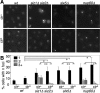
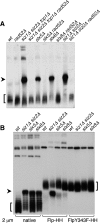
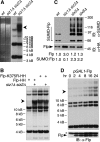
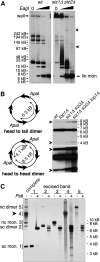
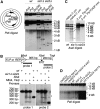
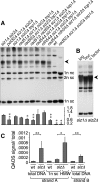
Many Saccharomyces cerevisiae mutants defective in the SUMO pathway accumulate elevated levels of the native 2 microm circle plasmid (2 microm). Here we show that accumulation of 2 microm in the SUMO pathway mutants siz1Delta siz2Delta, slx5Delta, and slx8Delta is associated with formation of an aberrant high-molecular-weight (HMW) form of 2 microm. Characterization of this species from siz1Delta siz2Delta showed that it contains tandem copies of the 2 mum sequence as well as single-stranded DNA. Accumulation of this species requires both the 2 microm-encoded Flp recombinase and the cellular homologous recombination repair (HRR) pathway. Importantly, reduced SUMO attachment to Flp is sufficient to induce formation of this species. Our data suggest a model in which Flp that cannot be sumoylated causes DNA damage, whose repair via HRR produces an intermediate that generates tandem copies of the 2 microm sequence. This intermediate may be a rolling circle formed via break-induced replication (BIR), because mutants defective in BIR contain reduced levels of the HMW form. This work also illustrates the importance of using cir(o) strains when studying mutants that affect the yeast SUMO pathway, to avoid confusing direct functions of the SUMO pathway with secondary effects of 2 microm amplification.
Figures
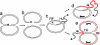
Similar articles
-
Misregulation of 2 microm circle copy number in a SUMO pathway mutant.Mol Cell Biol. 2005 May;25(10):4311-20. doi: 10.1128/MCB.25.10.4311-4320.2005. Mol Cell Biol. 2005. PMID: 15870299 Free PMC article.
-
RAD52-dependent and -independent homologous recombination initiated by Flp recombinase at a single FRT site flanked by direct repeats.Mol Gen Genet. 2000 Feb;263(1):73-80. doi: 10.1007/pl00008677. Mol Gen Genet. 2000. PMID: 10732675
-
Involvement of the inverted repeat of the yeast 2-micron plasmid in Flp site-specific and RAD52-dependent homologous recombination.Mol Gen Genet. 2000 Feb;263(1):81-9. doi: 10.1007/pl00008678. Mol Gen Genet. 2000. PMID: 10732676
-
The 2 micron plasmid of Saccharomyces cerevisiae: a miniaturized selfish genome with optimized functional competence.Plasmid. 2013 Jul;70(1):2-17. doi: 10.1016/j.plasmid.2013.03.001. Epub 2013 Mar 27. Plasmid. 2013. PMID: 23541845 Review.
-
Regulation of rDNA stability by sumoylation.DNA Repair (Amst). 2009 Apr 5;8(4):507-16. doi: 10.1016/j.dnarep.2009.01.015. Epub 2009 Mar 3. DNA Repair (Amst). 2009. PMID: 19261548 Review.
Cited by
-
Expression of Flp Protein in a Baculovirus/Insect Cell System for Biotechnological Applications.Protein J. 2017 Aug;36(4):332-342. doi: 10.1007/s10930-017-9724-z. Protein J. 2017. PMID: 28660316
-
Genetic evidence that polysumoylation bypasses the need for a SUMO-targeted Ub ligase.Genetics. 2011 Jan;187(1):73-87. doi: 10.1534/genetics.110.124347. Epub 2010 Nov 8. Genetics. 2011. PMID: 21059884 Free PMC article.
-
Genome maintenance in Saccharomyces cerevisiae: the role of SUMO and SUMO-targeted ubiquitin ligases.Nucleic Acids Res. 2017 Mar 17;45(5):2242-2261. doi: 10.1093/nar/gkw1369. Nucleic Acids Res. 2017. PMID: 28115630 Free PMC article. Review.
-
Expanding the scope of site-specific recombinases for genetic and metabolic engineering.Biotechnol Bioeng. 2014 Jan;111(1):1-15. doi: 10.1002/bit.25096. Epub 2013 Sep 13. Biotechnol Bioeng. 2014. PMID: 23982993 Free PMC article. Review.
-
The selfish yeast plasmid exploits a SWI/SNF-type chromatin remodeling complex for hitchhiking on chromosomes and ensuring high-fidelity propagation.PLoS Genet. 2023 Oct 9;19(10):e1010986. doi: 10.1371/journal.pgen.1010986. eCollection 2023 Oct. PLoS Genet. 2023. PMID: 37812641 Free PMC article.
References
-
- Adkins M. W., Tyler J. K. The histone chaperone Asf1p mediates global chromatin disassembly in vivo. J. Biol. Chem. 2004;279:52069–52074. - PubMed
-
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Smith J. A., Seidman J. G., Struhl K. Current Protocols in Molecular Biology. New York: Wiley-Interscience; 2000.
-
- Broach J. R., Volkert F. C. Circular DNA plasmids of yeasts. In: Broach J. R., Jones E. W., Pringle J. R., editors. The Molecular and Cellular Biology of the Yeast Saccharomyces. Vol. 1. Plainview, NY: Cold Spring Harbor Press; 1991. pp. 297–331.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

