Purification of SUMO conjugating enzymes and kinetic analysis of substrate conjugation
- PMID: 19107417
- PMCID: PMC2739043
- DOI: 10.1007/978-1-59745-566-4_11
Purification of SUMO conjugating enzymes and kinetic analysis of substrate conjugation
Abstract
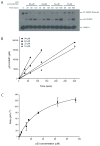
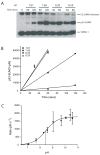
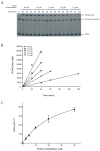
SUMO conjugation to protein substrates requires the concerted action of a dedicated E2 ubiquitin conjugation enzyme (Ubc9) and associated E3 ligases. Although Ubc9 can directly recognize and modify substrate lysine residues that occur within a consensus site for SUMO modification, E3 ligases can redirect specificity and enhance conjugation rates during SUMO conjugation in vitro and in vivo. In this chapter, we will describe methods utilized to purify SUMO conjugating enzymes and model substrates which can be used for analysis of SUMO conjugation in vitro. We will also describe methods to extract kinetic parameters during E3-dependent or E3-independent substrate conjugation.
Figures
Similar articles
-
Biochemical characterization of SUMO-conjugating enzymes by in vitro sumoylation assays.Methods Enzymol. 2019;618:167-185. doi: 10.1016/bs.mie.2018.12.025. Epub 2019 Feb 11. Methods Enzymol. 2019. PMID: 30850051
-
Role of an N-terminal site of Ubc9 in SUMO-1, -2, and -3 binding and conjugation.Biochemistry. 2003 Aug 26;42(33):9959-69. doi: 10.1021/bi0345283. Biochemistry. 2003. PMID: 12924945
-
Molecular mechanisms in SUMO conjugation.Biochem Soc Trans. 2020 Feb 28;48(1):123-135. doi: 10.1042/BST20190357. Biochem Soc Trans. 2020. PMID: 31872228 Review.
-
Purification and activity assays for Ubc9, the ubiquitin-conjugating enzyme for the small ubiquitin-like modifier SUMO.Methods Enzymol. 2005;398:74-87. doi: 10.1016/S0076-6879(05)98008-7. Methods Enzymol. 2005. PMID: 16275321
-
SUMO: getting it on.Biochem Soc Trans. 2007 Dec;35(Pt 6):1409-13. doi: 10.1042/BST0351409. Biochem Soc Trans. 2007. PMID: 18031233 Review.
Cited by
-
DNA-dependent SUMO modification of PARP-1.DNA Repair (Amst). 2013 Sep;12(9):761-73. doi: 10.1016/j.dnarep.2013.07.001. Epub 2013 Jul 18. DNA Repair (Amst). 2013. PMID: 23871147 Free PMC article.
-
Interactions of the Metalloregulatory Protein SloR from Streptococcus mutans with Its Metal Ion Effectors and DNA Binding Site.J Bacteriol. 2015 Nov;197(22):3601-15. doi: 10.1128/JB.00612-15. Epub 2015 Sep 8. J Bacteriol. 2015. PMID: 26350131 Free PMC article.
-
E2-mediated small ubiquitin-like modifier (SUMO) modification of thymine DNA glycosylase is efficient but not selective for the enzyme-product complex.J Biol Chem. 2014 May 30;289(22):15810-9. doi: 10.1074/jbc.M114.572081. Epub 2014 Apr 21. J Biol Chem. 2014. PMID: 24753249 Free PMC article.
-
Molecular mechanism of K65 acetylation-induced attenuation of Ubc9 and the NDSM interaction.Sci Rep. 2017 Dec 12;7(1):17391. doi: 10.1038/s41598-017-17465-0. Sci Rep. 2017. PMID: 29234076 Free PMC article.
-
Capturing a substrate in an activated RING E3/E2-SUMO complex.Nature. 2016 Aug 18;536(7616):304-8. doi: 10.1038/nature19071. Epub 2016 Aug 10. Nature. 2016. PMID: 27509863 Free PMC article.
References
-
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. - PubMed
-
- Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. - PubMed
-
- Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. - PubMed
-
- Laney JD, Hochstrasser M. Substrate targeting in the ubiquitin system. Cell. 1999;97:427–430. - PubMed
-
- Melchior F. SUMO--nonclassical ubiquitin. Annu Rev Cell Dev Biol. 2000;16:591–626. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

