Effects of CpG methylation on recognition of DNA by the tumour suppressor p53
- PMID: 19084536
- PMCID: PMC2666794
- DOI: 10.1016/j.jmb.2008.11.054
Effects of CpG methylation on recognition of DNA by the tumour suppressor p53
Abstract
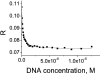
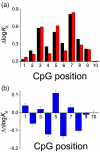
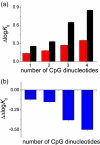
Methylation of DNA is one of the mechanisms controlling the expression landscape of the genome. Its pattern is altered in cancer and often results in the hypermethylation of the promoter regions and abnormal expression of tumour suppressor genes. Methylation of CpG dinucleotides located in the binding sites of transcription factors may contribute to the development of cancers by preventing their binding or altering their specificity. We studied the effects of CpG methylation on DNA recognition by the tumour suppressor p53, a transcription factor involved in the response to carcinogenic stress. p53 recognises a large number of DNA sequences, many of which contain CpG dinucleotides. We systematically substituted a CpG dinucleotide at each position in the consensus p53 DNA binding sequence and identified substitutions tolerated by p53. We compared the binding affinities of methylated versus non-methylated sequences by fluorescence anisotropy titration. We found that binding of p53 was not affected by cytosine methylation in a majority of cases. However, for a few sequences containing multiple CpG dinucleotides, such as sites in the RB and Met genes, methylation resulted in a four- to sixfold increase in binding of p53. This approach can be used to quantify the effects of CpG methylation on the DNA recognition by other DNA-binding proteins.
Figures

Similar articles
-
Unveiling the methylation status of CpG dinucleotides in the substituted segment of the human p53 knock-in (Hupki) mouse genome.Mol Carcinog. 2010 Dec;49(12):999-1006. doi: 10.1002/mc.20683. Mol Carcinog. 2010. PMID: 20945503 Free PMC article.
-
CpG methylation inactivates the transcriptional activity of the promoter of the human p53 tumor suppressor gene.Biochem Biophys Res Commun. 1997 Jun 18;235(2):403-6. doi: 10.1006/bbrc.1997.6796. Biochem Biophys Res Commun. 1997. PMID: 9199206
-
Methylation and deamination of CpGs generate p53-binding sites on a genomic scale.Trends Genet. 2009 Feb;25(2):63-6. doi: 10.1016/j.tig.2008.11.005. Epub 2008 Dec 26. Trends Genet. 2009. PMID: 19101055
-
Monitoring methylation changes in cancer.Adv Biochem Eng Biotechnol. 2007;104:1-11. doi: 10.1007/10_024. Adv Biochem Eng Biotechnol. 2007. PMID: 17290816 Review.
-
Aberrant DNA methylation in cancer: potential clinical interventions.Expert Rev Mol Med. 2002 Mar 4;4(4):1-17. doi: 10.1017/S1462399402004222. Expert Rev Mol Med. 2002. PMID: 14987388 Review.
Cited by
-
DNA methylation induces subtle mechanical alteration but significant chiral selectivity.Chem Commun (Camb). 2023 Dec 14;59(100):14855-14858. doi: 10.1039/d3cc05211g. Chem Commun (Camb). 2023. PMID: 38015496 Free PMC article.
-
Mechanisms of transcription factor selectivity.Trends Genet. 2010 Feb;26(2):75-83. doi: 10.1016/j.tig.2009.12.003. Epub 2010 Jan 13. Trends Genet. 2010. PMID: 20074831 Free PMC article. Review.
-
Local depletion of DNA methylation identifies a repressive p53 regulatory region in the NEK2 promoter.J Biol Chem. 2013 Dec 13;288(50):35940-51. doi: 10.1074/jbc.M113.523837. Epub 2013 Oct 25. J Biol Chem. 2013. PMID: 24163369 Free PMC article.
-
Conservation of DNA-binding specificity and oligomerisation properties within the p53 family.BMC Genomics. 2009 Dec 23;10:628. doi: 10.1186/1471-2164-10-628. BMC Genomics. 2009. PMID: 20030809 Free PMC article.
-
Distinct p53 genomic binding patterns in normal and cancer-derived human cells.Cell Cycle. 2011 Dec 15;10(24):4237-49. doi: 10.4161/cc.10.24.18383. Epub 2011 Dec 15. Cell Cycle. 2011. PMID: 22127205 Free PMC article.
References
-
- Robertson K.D. DNA methylation and human disease. Nat. Rev. Genet. 2005;6:597–610. - PubMed
-
- Brena R.M., Costello J.F. Genome–epigenome interactions in cancer. Hum. Mol. Genet. 2007;16(Spec. No. 1):R96–R105. - PubMed
-
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. - PubMed
-
- Watt F., Molloy P.L. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 1988;2:1136–1143. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

