Collapse transition in proteins
- PMID: 19081910
- PMCID: PMC2659394
- DOI: 10.1039/b813961j
Collapse transition in proteins
Abstract
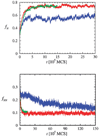
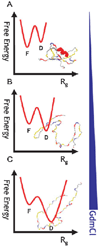
The coil-globule transition, a tenet of the physics of polymers, has been identified in recent years as an important unresolved aspect of the initial stages of the folding of proteins. We describe the basics of the collapse transition, starting with homopolymers and continuing with proteins. Studies of denatured-state collapse under equilibrium are then presented. An emphasis is placed on single-molecule fluorescence experiments, which are particularly useful for measuring properties of the denatured state even under conditions of coexistence with the folded state. Attempts to understand the dynamics of collapse, both theoretically and experimentally, are then described. Only an upper limit for the rate of collapse has been obtained so far. Improvements in experimental and theoretical methodology are likely to continue to push our understanding of the importance of the denatured-state thermodynamics and dynamics for protein folding in the coming years.
Figures
Similar articles
-
Protein folding, protein collapse, and tanford's transfer model: lessons from single-molecule FRET.J Am Chem Soc. 2009 Mar 4;131(8):2942-7. doi: 10.1021/ja808305u. J Am Chem Soc. 2009. PMID: 19239269 Free PMC article.
-
How, when and why proteins collapse: the relation to folding.Curr Opin Struct Biol. 2012 Feb;22(1):14-20. doi: 10.1016/j.sbi.2011.10.005. Epub 2011 Nov 19. Curr Opin Struct Biol. 2012. PMID: 22104965 Free PMC article. Review.
-
Coil-globule transition in the denatured state of a small protein.Proc Natl Acad Sci U S A. 2006 Aug 1;103(31):11539-43. doi: 10.1073/pnas.0601395103. Epub 2006 Jul 20. Proc Natl Acad Sci U S A. 2006. PMID: 16857738 Free PMC article.
-
Macromolecular crowding: chemistry and physics meet biology (Ascona, Switzerland, 10-14 June 2012).Phys Biol. 2013 Aug;10(4):040301. doi: 10.1088/1478-3975/10/4/040301. Epub 2013 Aug 2. Phys Biol. 2013. PMID: 23912807
-
Role of solvation effects in protein denaturation: from thermodynamics to single molecules and back.Annu Rev Phys Chem. 2011;62:257-77. doi: 10.1146/annurev-physchem-032210-103531. Annu Rev Phys Chem. 2011. PMID: 21219136 Free PMC article. Review.
Cited by
-
Dramatic Shape Changes Occur as Cytochrome c Folds.J Phys Chem B. 2020 Sep 24;124(38):8240-8248. doi: 10.1021/acs.jpcb.0c05802. Epub 2020 Sep 9. J Phys Chem B. 2020. PMID: 32840372 Free PMC article.
-
Slow unfolded-state structuring in Acyl-CoA binding protein folding revealed by simulation and experiment.J Am Chem Soc. 2012 Aug 1;134(30):12565-77. doi: 10.1021/ja302528z. Epub 2012 Jul 19. J Am Chem Soc. 2012. PMID: 22747188 Free PMC article.
-
Conformations of a Metastable SH3 Domain Characterized by smFRET and an Excluded-Volume Polymer Model.Biophys J. 2016 Apr 12;110(7):1510-1522. doi: 10.1016/j.bpj.2016.02.033. Biophys J. 2016. PMID: 27074677 Free PMC article.
-
Decoupling of size and shape fluctuations in heteropolymeric sequences reconciles discrepancies in SAXS vs. FRET measurements.Proc Natl Acad Sci U S A. 2017 Aug 1;114(31):E6342-E6351. doi: 10.1073/pnas.1704692114. Epub 2017 Jul 17. Proc Natl Acad Sci U S A. 2017. PMID: 28716919 Free PMC article.
-
Integrating single-molecule spectroscopy and simulations for the study of intrinsically disordered proteins.Methods. 2021 Sep;193:116-135. doi: 10.1016/j.ymeth.2021.03.018. Epub 2021 Apr 6. Methods. 2021. PMID: 33831596 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

