AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations
- PMID: 19070574
- PMCID: PMC2713603
- DOI: 10.1016/j.cell.2008.09.062
AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations
Abstract
Chromosomal translocation requires formation of paired double-strand DNA breaks (DSBs) on heterologous chromosomes. One of the most well characterized oncogenic translocations juxtaposes c-myc and the immunoglobulin heavy-chain locus (IgH) and is found in Burkitt's lymphomas in humans and plasmacytomas in mice. DNA breaks in IgH leading to c-myc/IgH translocations are created by activation-induced cytidine deaminase (AID) during antibody class switch recombination or somatic hypermutation. However, the source of DNA breaks at c-myc is not known. Here, we provide evidence for the c-myc promoter region being required in targeting AID-mediated DNA damage to produce DSBs in c-myc that lead to c-myc/IgH translocations in primary B lymphocytes. Thus, in addition to producing somatic mutations and DNA breaks in antibody genes, AID is also responsible for the DNA lesions in oncogenes that are required for their translocation.
Figures


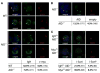

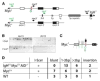


Comment in
-
Collateral damage from antigen receptor gene diversification.Cell. 2008 Dec 12;135(6):1009-12. doi: 10.1016/j.cell.2008.11.024. Cell. 2008. PMID: 19070571 Review.
Similar articles
-
Mechanisms promoting translocations in editing and switching peripheral B cells.Nature. 2009 Jul 9;460(7252):231-6. doi: 10.1038/nature08159. Nature. 2009. PMID: 19587764 Free PMC article.
-
Role of the translocation partner in protection against AID-dependent chromosomal translocations.Proc Natl Acad Sci U S A. 2010 Jan 5;107(1):187-92. doi: 10.1073/pnas.0908946107. Epub 2009 Dec 4. Proc Natl Acad Sci U S A. 2010. PMID: 19966290 Free PMC article.
-
Role of genomic instability and p53 in AID-induced c-myc-Igh translocations.Nature. 2006 Mar 2;440(7080):105-9. doi: 10.1038/nature04495. Epub 2006 Jan 8. Nature. 2006. PMID: 16400328 Free PMC article.
-
AID and Igh switch region-Myc chromosomal translocations.DNA Repair (Amst). 2006 Sep 8;5(9-10):1259-64. doi: 10.1016/j.dnarep.2006.05.019. Epub 2006 Jun 19. DNA Repair (Amst). 2006. PMID: 16784901 Review.
-
Restricting activation-induced cytidine deaminase tumorigenic activity in B lymphocytes.Immunology. 2009 Mar;126(3):316-28. doi: 10.1111/j.1365-2567.2008.03050.x. Immunology. 2009. PMID: 19302140 Free PMC article. Review.
Cited by
-
Mouse model of endemic Burkitt translocations reveals the long-range boundaries of Ig-mediated oncogene deregulation.Proc Natl Acad Sci U S A. 2012 Jul 3;109(27):10972-7. doi: 10.1073/pnas.1200106109. Epub 2012 Jun 18. Proc Natl Acad Sci U S A. 2012. PMID: 22711821 Free PMC article.
-
AID in Chronic Lymphocytic Leukemia: Induction and Action During Disease Progression.Front Oncol. 2021 May 10;11:634383. doi: 10.3389/fonc.2021.634383. eCollection 2021. Front Oncol. 2021. PMID: 34041018 Free PMC article. Review.
-
Oncogenic Enhancers in Leukemia.Blood Cancer Discov. 2024 Sep 3;5(5):303-317. doi: 10.1158/2643-3230.BCD-23-0211. Blood Cancer Discov. 2024. PMID: 39093124 Review.
-
Mouse models of diffuse large B cell lymphoma.Front Immunol. 2023 Dec 6;14:1313371. doi: 10.3389/fimmu.2023.1313371. eCollection 2023. Front Immunol. 2023. PMID: 38124747 Free PMC article. Review.
-
The dangers of transcription.Cell. 2009 Dec 11;139(6):1047-9. doi: 10.1016/j.cell.2009.11.037. Cell. 2009. PMID: 20005797 Free PMC article.
References
-
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. - PubMed
-
- Belotserkovskii BP, De Silva E, Tornaletti S, Wang G, Vasquez KM, Hanawalt PC. A triplex-forming sequence from the human c-MYC promoter interferes with DNA transcription. J Biol Chem. 2007;282:32433–32441. - PubMed
-
- Boles TC, Hogan ME. DNA structure equilibria in the human c-myc gene. Biochemistry. 1987;26:367–376. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI037526/AI/NIAID NIH HHS/United States
- R01 AI037526-15/AI/NIAID NIH HHS/United States
- BBS/E/B/0000C163/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/F012217/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- ImNIH/Intramural NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

