Identification, structure, and functional requirement of the Mediator submodule Med7N/31
- PMID: 19057509
- PMCID: PMC2633081
- DOI: 10.1038/emboj.2008.254
Identification, structure, and functional requirement of the Mediator submodule Med7N/31
Abstract
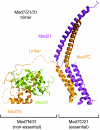
Mediator is a modular multiprotein complex required for regulated transcription by RNA polymerase (Pol) II. Here, we show that the middle module of the Mediator core contains a submodule of unique structure and function that comprises the N-terminal part of subunit Med7 (Med7N) and the highly conserved subunit Med31 (Soh1). The Med7N/31 submodule shows a conserved novel fold, with two proline-rich stretches in Med7N wrapping around the right-handed four-helix bundle of Med31. In vitro, Med7N/31 is required for activated transcription and can act in trans when added exogenously. In vivo, Med7N/31 has a predominantly positive function on the expression of a specific subset of genes, including genes involved in methionine metabolism and iron transport. Comparative phenotyping and transcriptome profiling identify specific and overlapping functions of different Mediator submodules.
Figures



Similar articles
-
A conserved mediator hinge revealed in the structure of the MED7.MED21 (Med7.Srb7) heterodimer.J Biol Chem. 2005 May 6;280(18):18171-8. doi: 10.1074/jbc.M413466200. Epub 2005 Feb 14. J Biol Chem. 2005. PMID: 15710619
-
The Nuclear Pore-Associated TREX-2 Complex Employs Mediator to Regulate Gene Expression.Cell. 2015 Aug 27;162(5):1016-28. doi: 10.1016/j.cell.2015.07.059. Cell. 2015. PMID: 26317468 Free PMC article.
-
The structural and functional organization of the yeast mediator complex.J Biol Chem. 2001 Nov 9;276(45):42003-10. doi: 10.1074/jbc.M105961200. Epub 2001 Sep 12. J Biol Chem. 2001. PMID: 11555651
-
Interactions between subunits of Drosophila Mediator and activator proteins.Trends Biochem Sci. 2005 May;30(5):245-9. doi: 10.1016/j.tibs.2005.03.010. Trends Biochem Sci. 2005. PMID: 15896742 Review.
-
The yeast Mediator complex and its regulation.Trends Biochem Sci. 2005 May;30(5):240-4. doi: 10.1016/j.tibs.2005.03.008. Trends Biochem Sci. 2005. PMID: 15896741 Review.
Cited by
-
Mechanisms of Mediator complex action in transcriptional activation.Cell Mol Life Sci. 2013 Aug;70(15):2743-56. doi: 10.1007/s00018-013-1265-9. Epub 2013 Jan 30. Cell Mol Life Sci. 2013. PMID: 23361037 Free PMC article. Review.
-
Preparation and topology of the Mediator middle module.Nucleic Acids Res. 2010 Jun;38(10):3186-95. doi: 10.1093/nar/gkq029. Epub 2010 Jan 31. Nucleic Acids Res. 2010. PMID: 20123732 Free PMC article.
-
The Mediator complex in plants: structure, phylogeny, and expression profiling of representative genes in a dicot (Arabidopsis) and a monocot (rice) during reproduction and abiotic stress.Plant Physiol. 2011 Dec;157(4):1609-27. doi: 10.1104/pp.111.188300. Epub 2011 Oct 21. Plant Physiol. 2011. PMID: 22021418 Free PMC article.
-
The Mediator complex and transcription regulation.Crit Rev Biochem Mol Biol. 2013 Nov-Dec;48(6):575-608. doi: 10.3109/10409238.2013.840259. Epub 2013 Oct 3. Crit Rev Biochem Mol Biol. 2013. PMID: 24088064 Free PMC article. Review.
-
Systematic analysis of the expression profiles and prognostic significance of the MED gene family in renal clear cell carcinoma.Oncol Lett. 2024 Jun 26;28(2):398. doi: 10.3892/ol.2024.14531. eCollection 2024 Aug. Oncol Lett. 2024. PMID: 38979551 Free PMC article.
References
-
- Adzhubei AA, Sternberg MJ (1993) Left-handed polyproline II helices commonly occur in globular proteins. J Mol Biol 229: 472–493 - PubMed
-
- Alexa A, Rahnenfuhrer J, Lengauer T (2006) Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22: 1600–1607 - PubMed
-
- Asturias FJ, Jiang YW, Myers LC, Gustafsson CM, Kornberg RD (1999) Conserved structures of Mediator and RNA polymerase II holoenzyme. Science 283: 985–987 - PubMed
-
- Bäckström S, Elfving N, Nilsson R, Wingsle G, Björklund S (2007) Purification of a plant Mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol Cell 26: 717–729 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

