Multiple nucleic acid binding sites and intrinsic disorder of severe acute respiratory syndrome coronavirus nucleocapsid protein: implications for ribonucleocapsid protein packaging
- PMID: 19052082
- PMCID: PMC2643731
- DOI: 10.1128/JVI.02001-08
Multiple nucleic acid binding sites and intrinsic disorder of severe acute respiratory syndrome coronavirus nucleocapsid protein: implications for ribonucleocapsid protein packaging
Abstract
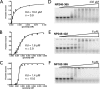
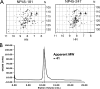
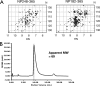
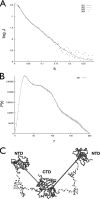
The nucleocapsid protein (N) of the severe acute respiratory syndrome coronavirus (SARS-CoV) packages the viral genomic RNA and is crucial for viability. However, the RNA-binding mechanism is poorly understood. We have shown previously that the N protein contains two structural domains--the N-terminal domain (NTD; residues 45 to 181) and the C-terminal dimerization domain (CTD; residues 248 to 365)--flanked by long stretches of disordered regions accounting for almost half of the entire sequence. Small-angle X-ray scattering data show that the protein is in an extended conformation and that the two structural domains of the SARS-CoV N protein are far apart. Both the NTD and the CTD have been shown to bind RNA. Here we show that all disordered regions are also capable of binding to RNA. Constructs containing multiple RNA-binding regions showed Hill coefficients greater than 1, suggesting that the N protein binds to RNA cooperatively. The effect can be explained by the "coupled-allostery" model, devised to explain the allosteric effect in a multidomain regulatory system. Although the N proteins of different coronaviruses share very low sequence homology, the physicochemical features described above may be conserved across different groups of Coronaviridae. The current results underscore the important roles of multisite nucleic acid binding and intrinsic disorder in N protein function and RNP packaging.
Figures

Similar articles
-
Solution structure of the c-terminal dimerization domain of SARS coronavirus nucleocapsid protein solved by the SAIL-NMR method.J Mol Biol. 2008 Jul 18;380(4):608-22. doi: 10.1016/j.jmb.2007.11.093. Epub 2007 Dec 5. J Mol Biol. 2008. PMID: 18561946 Free PMC article.
-
The SARS coronavirus nucleocapsid protein--forms and functions.Antiviral Res. 2014 Mar;103:39-50. doi: 10.1016/j.antiviral.2013.12.009. Epub 2014 Jan 11. Antiviral Res. 2014. PMID: 24418573 Free PMC article. Review.
-
Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA.J Mol Biol. 2007 May 11;368(4):1075-86. doi: 10.1016/j.jmb.2007.02.069. Epub 2007 Mar 2. J Mol Biol. 2007. PMID: 17379242 Free PMC article.
-
Carboxyl terminus of severe acute respiratory syndrome coronavirus nucleocapsid protein: self-association analysis and nucleic acid binding characterization.Biochemistry. 2006 Oct 3;45(39):11827-35. doi: 10.1021/bi0609319. Biochemistry. 2006. PMID: 17002283
-
The SARS-CoV nucleocapsid protein: a protein with multifarious activities.Infect Genet Evol. 2008 Jul;8(4):397-405. doi: 10.1016/j.meegid.2007.07.004. Epub 2007 Jul 20. Infect Genet Evol. 2008. PMID: 17881296 Free PMC article. Review.
Cited by
-
Inhibition of SARS-CoV-2 replication by a ssDNA aptamer targeting the nucleocapsid protein.Microbiol Spectr. 2024 Apr 2;12(4):e0341023. doi: 10.1128/spectrum.03410-23. Epub 2024 Feb 20. Microbiol Spectr. 2024. PMID: 38376366 Free PMC article.
-
Molecular mechanisms of the novel coronavirus SARS-CoV-2 and potential anti-COVID19 pharmacological targets since the outbreak of the pandemic.Food Chem Toxicol. 2020 Dec;146:111805. doi: 10.1016/j.fct.2020.111805. Epub 2020 Oct 8. Food Chem Toxicol. 2020. PMID: 33038452 Free PMC article. Review.
-
SARS-CoV-2 Nucleocapsid Protein Targets a Conserved Surface Groove of the NTF2-like Domain of G3BP1.J Mol Biol. 2022 May 15;434(9):167516. doi: 10.1016/j.jmb.2022.167516. Epub 2022 Feb 28. J Mol Biol. 2022. PMID: 35240128 Free PMC article.
-
The RNA binding of protein A from Wuhan nodavirus is mediated by mitochondrial membrane lipids.Virology. 2014 Aug;462-463:1-13. doi: 10.1016/j.virol.2014.05.022. Epub 2014 Jun 13. Virology. 2014. PMID: 25092456 Free PMC article.
-
A conserved oligomerization domain in the disordered linker of coronavirus nucleocapsid proteins.Sci Adv. 2023 Apr 5;9(14):eadg6473. doi: 10.1126/sciadv.adg6473. Epub 2023 Apr 5. Sci Adv. 2023. PMID: 37018390 Free PMC article.
References
-
- Chang, C. K., S. C. Sue, T. H. Yu, C. M. Hsieh, C. K. Tsai, Y. C. Chiang, S. J. Lee, H. H. Hsiao, W. J. Wu, C. F. Chang, and T. H. Huang. 2005. The dimer interface of the SARS coronavirus nucleocapsid protein adapts a porcine respiratory and reproductive syndrome virus-like structure. FEBS Lett. 5795663-5668. - PMC - PubMed
-
- Chiti, F., M. Stefani, N. Taddei, G. Ramponi, and C. M. Dobson. 2003. Rationalization of the effects of mutations on peptide and protein aggregation rates. Nature 424805-808. - PubMed
-
- Cuff, J. A., and G. J. Barton. 2000. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins 40502-511. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

