Highly dissociative and concerted mechanism for the nicotinamide cleavage reaction in Sir2Tm enzyme suggested by ab initio QM/MM molecular dynamics simulations
- PMID: 19049465
- PMCID: PMC2627508
- DOI: 10.1021/ja807269j
Highly dissociative and concerted mechanism for the nicotinamide cleavage reaction in Sir2Tm enzyme suggested by ab initio QM/MM molecular dynamics simulations
Abstract
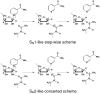
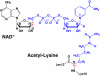
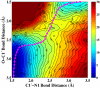
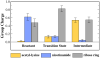
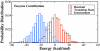
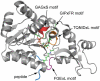
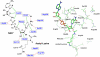
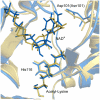
Sir2 enzymes catalyze the NAD+-dependent protein deacetylation and play critical roles in epigenetics, cell death, and lifespan regulation. In spite of a current flurry of experimental studies, the catalytic mechanism for this unique and important class of enzymes remains elusive. Employing on-the-fly Born-Oppenheimer molecular dynamics simulations with the B3LYP/6-31G(d) QM/MM potential and the umbrella sampling method, we have characterized the initial step of the Sir2Tm-catalyzed reaction, which is also the most controversial portion of its mechanism. Our results indicate that the nicotinamide cleavage reaction employs a highly dissociative and concerted displacement mechanism: the cleavage of the glycosidic bond is facilitated by the nucleophilic participation of the acetyl-lysine, and the dissociative transition state has a significant oxocarbenium ion character. During this step of the reaction, the Sir2Tm enzyme strongly stabilizes the covalent O-alkylamidate intermediate whereas its effect on the transition state is quite minimal. In addition, functional roles of key residues and motifs have been elucidated. This work further demonstrates the feasibility and applicability of the state-of-the-art ab initio QM/MM molecular dynamics approach in simulating enzyme reactions.
Figures


Similar articles
-
Catalytic reaction mechanism of acetylcholinesterase determined by Born-Oppenheimer ab initio QM/MM molecular dynamics simulations.J Phys Chem B. 2010 Jul 8;114(26):8817-25. doi: 10.1021/jp104258d. J Phys Chem B. 2010. PMID: 20550161 Free PMC article.
-
Investigation of the catalytic mechanism of Sir2 enzyme with QM/MM approach: SN1 vs SN2?J Phys Chem B. 2010 Sep 16;114(36):11927-33. doi: 10.1021/jp1054183. J Phys Chem B. 2010. PMID: 20726530
-
Structural insights into intermediate steps in the Sir2 deacetylation reaction.Structure. 2008 Sep 10;16(9):1368-77. doi: 10.1016/j.str.2008.05.015. Structure. 2008. PMID: 18786399 Free PMC article.
-
SIR2: the biochemical mechanism of NAD(+)-dependent protein deacetylation and ADP-ribosyl enzyme intermediates.Curr Med Chem. 2004 Apr;11(7):807-26. doi: 10.2174/0929867043455675. Curr Med Chem. 2004. PMID: 15078167 Review.
-
Mechanism-based modulator discovery for sirtuin-catalyzed deacetylation reaction.Mini Rev Med Chem. 2013 Jan;13(1):132-54. Mini Rev Med Chem. 2013. PMID: 22876953 Review.
Cited by
-
Catalytic reaction mechanism of acetylcholinesterase determined by Born-Oppenheimer ab initio QM/MM molecular dynamics simulations.J Phys Chem B. 2010 Jul 8;114(26):8817-25. doi: 10.1021/jp104258d. J Phys Chem B. 2010. PMID: 20550161 Free PMC article.
-
Molecular Dynamics Simulations with Quantum Mechanics/Molecular Mechanics and Adaptive Neural Networks.J Chem Theory Comput. 2018 Mar 13;14(3):1442-1455. doi: 10.1021/acs.jctc.7b01195. Epub 2018 Feb 26. J Chem Theory Comput. 2018. PMID: 29438614 Free PMC article.
-
How is acetylcholinesterase phosphonylated by soman? An ab initio QM/MM molecular dynamics study.J Phys Chem A. 2014 Oct 2;118(39):9132-9. doi: 10.1021/jp502712d. Epub 2014 May 9. J Phys Chem A. 2014. PMID: 24786171 Free PMC article.
-
Thiol versus hydroxamate as zinc binding group in HDAC inhibition: An ab initio QM/MM molecular dynamics study.J Comput Chem. 2015 Nov 15;36(30):2228-35. doi: 10.1002/jcc.24203. Epub 2015 Oct 9. J Comput Chem. 2015. PMID: 26452222 Free PMC article.
-
Transition state of ADP-ribosylation of acetyllysine catalyzed by Archaeoglobus fulgidus Sir2 determined by kinetic isotope effects and computational approaches.J Am Chem Soc. 2010 Sep 8;132(35):12286-98. doi: 10.1021/ja910342d. J Am Chem Soc. 2010. PMID: 20718419 Free PMC article.
References
-
- Blander G, Guarente L. Annu. Rev. Biochem. 2004;73:417–435. - PubMed
-
- Marmorstein R. Biochem. Soc. Trans. 2004;32:904–909. - PubMed
-
- Denu JM. Curr. Opin. Chem. biol. 2005;9:431–440. - PubMed
-
- Sauve AA, Wolberger C, Schramm VL, Boeke JD. Annu. Rev. Biochem. 2006;75:435–465. - PubMed
-
- Braunstein M, Rose AB, Holmes SG, Allis CD, Broach JR. Genes Dev. 1993;7:592–604. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

