Neurofibromin 1 (NF1) defects are common in human ovarian serous carcinomas and co-occur with TP53 mutations
- PMID: 19048115
- PMCID: PMC2586687
- DOI: 10.1593/neo.08784
Neurofibromin 1 (NF1) defects are common in human ovarian serous carcinomas and co-occur with TP53 mutations
Abstract
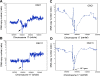
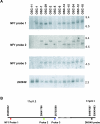
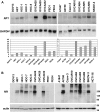
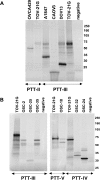
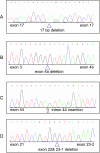
Ovarian serous carcinoma (OSC) is the most common and lethal histologic type of ovarian epithelial malignancy. Mutations of TP53 and dysfunction of the Brca1 and/or Brca2 tumor-suppressor proteins have been implicated in the molecular pathogenesis of a large fraction of OSCs, but frequent somatic mutations in other well-established tumor-suppressor genes have not been identified. Using a genome-wide screen of DNA copy number alterations in 36 primary OSCs, we identified two tumors with apparent homozygous deletions of the NF1 gene. Subsequently, 18 ovarian carcinoma-derived cell lines and 41 primary OSCs were evaluated for NF1 alterations. Markedly reduced or absent expression of Nf1 protein was observed in 6 of the 18 cell lines, and using the protein truncation test and sequencing of cDNA and genomic DNA, NF1 mutations resulting in deletion of exons and/or aberrant splicing of NF1 transcripts were detected in 5 of the 6 cell lines with loss of NF1 expression. Similarly, NF1 alterations including homozygous deletions and splicing mutations were identified in 9 (22%) of 41 primary OSCs. As expected, tumors and cell lines with NF1 defects lacked mutations in KRAS or BRAF but showed Ras pathway activation based on immunohistochemical detection of phosphorylated MAPK (primary tumors) or increased levels of GTP-bound Ras (cell lines). The TP53 tumor-suppressor gene was mutated in all OSCs with documented NF1 mutation, suggesting that the pathways regulated by these two tumor-suppressor proteins often cooperate in the development of ovarian carcinomas with serous differentiation.
Figures

Similar articles
-
TP53 mutations are common in all subtypes of epithelial ovarian cancer and occur concomitantly with KRAS mutations in the mucinous type.Exp Mol Pathol. 2013 Oct;95(2):235-41. doi: 10.1016/j.yexmp.2013.08.004. Epub 2013 Aug 18. Exp Mol Pathol. 2013. PMID: 23965232
-
Clinical and Molecular Characteristics of NF1-Mutant Lung Cancer.Clin Cancer Res. 2016 Jul 1;22(13):3148-56. doi: 10.1158/1078-0432.CCR-15-2377. Epub 2016 Feb 9. Clin Cancer Res. 2016. PMID: 26861459 Free PMC article.
-
Murine Oviductal High-Grade Serous Carcinomas Mirror the Genomic Alterations, Gene Expression Profiles, and Immune Microenvironment of Their Human Counterparts.Cancer Res. 2020 Feb 15;80(4):877-889. doi: 10.1158/0008-5472.CAN-19-2558. Epub 2019 Dec 5. Cancer Res. 2020. PMID: 31806642 Free PMC article.
-
[Pathogenic alterations within the neurofibromin gene in various cancers].Magy Onkol. 2017 Dec 18;61(4):327-336. Epub 2017 Sep 5. Magy Onkol. 2017. PMID: 29257151 Review. Hungarian.
-
A RASopathy gene commonly mutated in cancer: the neurofibromatosis type 1 tumour suppressor.Nat Rev Cancer. 2015 May;15(5):290-301. doi: 10.1038/nrc3911. Epub 2015 Apr 16. Nat Rev Cancer. 2015. PMID: 25877329 Free PMC article. Review.
Cited by
-
Copy number analysis identifies novel interactions between genomic loci in ovarian cancer.PLoS One. 2010 Sep 10;5(9):e11408. doi: 10.1371/journal.pone.0011408. PLoS One. 2010. PMID: 20844748 Free PMC article.
-
Clinical perspectives of BET inhibition in ovarian cancer.Cell Oncol (Dordr). 2021 Apr;44(2):237-249. doi: 10.1007/s13402-020-00578-6. Epub 2021 Jan 19. Cell Oncol (Dordr). 2021. PMID: 33469840 Review.
-
Genomic Alterations Correlated to Trastuzumab Resistance and Clinical Outcomes in HER2+/HR- Breast Cancers of Patients Living in Northwestern China.J Cancer. 2024 Jun 17;15(14):4467-4476. doi: 10.7150/jca.84832. eCollection 2024. J Cancer. 2024. PMID: 39006074 Free PMC article.
-
Multiple breast cancer cell-lines derived from a single tumor differ in their molecular characteristics and tumorigenic potential.PLoS One. 2013;8(1):e55145. doi: 10.1371/journal.pone.0055145. Epub 2013 Jan 25. PLoS One. 2013. PMID: 23372829 Free PMC article.
-
The neurofibromin recruitment factor Spred1 binds to the GAP related domain without affecting Ras inactivation.Proc Natl Acad Sci U S A. 2016 Jul 5;113(27):7497-502. doi: 10.1073/pnas.1607298113. Epub 2016 Jun 16. Proc Natl Acad Sci U S A. 2016. PMID: 27313208 Free PMC article.
References
-
- Seidman JD, Horkayne-Szakaly I, Haiba M, Boice CR, Kurman RJ, Ronnett BM. The histologic type and stage distribution of ovarian carcinomas of surface epithelial origin. Int J Gynecol Pathol. 2004;23:41–44. - PubMed
-
- Hilton JL, Geisler JP, Rathe JA, Hattermann-Zogg MA, DeYoung B, Buller RE. Inactivation of BRCA1 and BRCA2 in ovarian cancer. J Natl Cancer Inst. 2002;94:1396–1406. - PubMed
-
- Wang C, Horiuchi A, Imai T, Ohira S, Itoh K, Nikaido T, Katsuyama Y, Konishi I. Expression of BRCA1 protein in benign, borderline, and malignant epithelial ovarian neoplasms and its relationship to methylation and allelic loss of the BRCA1 gene. J Pathol. 2004;202:215–223. - PubMed
-
- Schuijer M, Berns EM. TP53 and ovarian cancer. Hum Mutat. 2003;21:285–291. - PubMed
-
- Pal T, Permuth-Wey J, Betts JA, Krischer JP, Fiorica J, Arango H, LaPolla J, Hoffman M, Martino MA, Wakeley K, et al. BRCA1 and BRCA2 mutations account for a large proportion of ovarian carcinoma cases. Cancer. 2005;104:2807–2816. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
