Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA
- PMID: 19043829
- PMCID: PMC2743566
- DOI: 10.1038/nature07413
Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA
Abstract
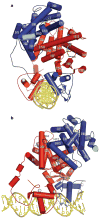
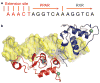
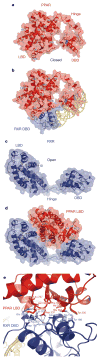
Nuclear receptors are multi-domain transcription factors that bind to DNA elements from which they regulate gene expression. The peroxisome proliferator-activated receptors (PPARs) form heterodimers with the retinoid X receptor (RXR), and PPAR-gamma has been intensively studied as a drug target because of its link to insulin sensitization. Previous structural studies have focused on isolated DNA or ligand-binding segments, with no demonstration of how multiple domains cooperate to modulate receptor properties. Here we present structures of intact PPAR-gamma and RXR-alpha as a heterodimer bound to DNA, ligands and coactivator peptides. PPAR-gamma and RXR-alpha form a non-symmetric complex, allowing the ligand-binding domain (LBD) of PPAR-gamma to contact multiple domains in both proteins. Three interfaces link PPAR-gamma and RXR-alpha, including some that are DNA dependent. The PPAR-gamma LBD cooperates with both DNA-binding domains (DBDs) to enhance response-element binding. The A/B segments are highly dynamic, lacking folded substructures despite their gene-activation properties.
Figures

Similar articles
-
Allosteric Pathways in the PPARγ-RXRα nuclear receptor complex.Sci Rep. 2016 Jan 29;6:19940. doi: 10.1038/srep19940. Sci Rep. 2016. PMID: 26823026 Free PMC article.
-
Low-resolution molecular models reveal the oligomeric state of the PPAR and the conformational organization of its domains in solution.PLoS One. 2012;7(2):e31852. doi: 10.1371/journal.pone.0031852. Epub 2012 Feb 21. PLoS One. 2012. PMID: 22363753 Free PMC article.
-
Synergistic Regulation of Coregulator/Nuclear Receptor Interaction by Ligand and DNA.Structure. 2017 Oct 3;25(10):1506-1518.e4. doi: 10.1016/j.str.2017.07.019. Epub 2017 Sep 7. Structure. 2017. PMID: 28890360 Free PMC article.
-
Modulation of RXR function through ligand design.Biochim Biophys Acta. 2012 Jan;1821(1):57-69. doi: 10.1016/j.bbalip.2011.04.003. Epub 2011 Apr 16. Biochim Biophys Acta. 2012. PMID: 21515403 Review.
-
Peroxisome proliferator-activated receptor gamma in malignant diseases.Crit Rev Oncol Hematol. 2006 Apr;58(1):1-14. doi: 10.1016/j.critrevonc.2005.08.011. Epub 2006 Jan 18. Crit Rev Oncol Hematol. 2006. PMID: 16388966 Review.
Cited by
-
Nuclear hormone receptor functions in keratinocyte and melanocyte homeostasis, epidermal carcinogenesis and melanomagenesis.FEBS Lett. 2013 Mar 18;587(6):529-41. doi: 10.1016/j.febslet.2013.01.041. Epub 2013 Feb 5. FEBS Lett. 2013. PMID: 23395795 Free PMC article. Review.
-
Mechanisms of action, chemical characteristics, and model systems of obesogens.BMC Biomed Eng. 2020 Apr 30;2:6. doi: 10.1186/s42490-020-00040-6. eCollection 2020. BMC Biomed Eng. 2020. PMID: 32903358 Free PMC article. Review.
-
The nuclear receptor signalling scaffold: insights from full-length structures.EMBO J. 2012 Jan 18;31(2):251-3. doi: 10.1038/emboj.2011.475. Epub 2012 Jan 18. EMBO J. 2012. PMID: 22252143 Free PMC article.
-
Prospective functional classification of all possible missense variants in PPARG.Nat Genet. 2016 Dec;48(12):1570-1575. doi: 10.1038/ng.3700. Epub 2016 Oct 17. Nat Genet. 2016. PMID: 27749844 Free PMC article.
-
Identification of a binding motif specific to HNF4 by comparative analysis of multiple nuclear receptors.Nucleic Acids Res. 2012 Jul;40(12):5343-56. doi: 10.1093/nar/gks190. Epub 2012 Mar 1. Nucleic Acids Res. 2012. PMID: 22383578 Free PMC article.
References
-
- Nagy L, Schwabe JW. Mechanism of the nuclear receptor molecular switch. Trends Biochem Sci. 2004;29:317–324. - PubMed
-
- Yin L, et al. Rev-erbα, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–1789. - PubMed
-
- Bain DL, Heneghan AF, Connaghan-Jones KD, Miura MT. Nuclear receptor structure: implications for function. Annu Rev Physiol. 2007;69:201–220. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

