Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer
- PMID: 19008416
- PMCID: PMC2684823
- DOI: 10.1126/science.1165395
Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer
Abstract
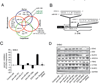
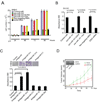
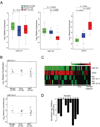
Enhancer of zeste homolog 2 (EZH2) is a mammalian histone methyltransferase that contributes to the epigenetic silencing of target genes and regulates the survival and metastasis of cancer cells. EZH2 is overexpressed in aggressive solid tumors by mechanisms that remain unclear. Here we show that the expression and function of EZH2 in cancer cell lines are inhibited by microRNA-101 (miR-101). Analysis of human prostate tumors revealed that miR-101 expression decreases during cancer progression, paralleling an increase in EZH2 expression. One or both of the two genomic loci encoding miR-101 were somatically lost in 37.5% of clinically localized prostate cancer cells (6 of 16) and 66.7% of metastatic disease cells (22 of 33). We propose that the genomic loss of miR-101 in cancer leads to overexpression of EZH2 and concomitant dysregulation of epigenetic pathways, resulting in cancer progression.
Figures

Similar articles
-
Polycomb protein EZH2 regulates tumor invasion via the transcriptional repression of the metastasis suppressor RKIP in breast and prostate cancer.Cancer Res. 2012 Jun 15;72(12):3091-104. doi: 10.1158/0008-5472.CAN-11-3546. Epub 2012 Apr 13. Cancer Res. 2012. PMID: 22505648
-
Polycomb protein EZH2 suppresses apoptosis by silencing the proapoptotic miR-31.Cell Death Dis. 2014 Oct 23;5(10):e1486. doi: 10.1038/cddis.2014.454. Cell Death Dis. 2014. PMID: 25341040 Free PMC article.
-
Repression of E-cadherin by the polycomb group protein EZH2 in cancer.Oncogene. 2008 Dec 11;27(58):7274-84. doi: 10.1038/onc.2008.333. Epub 2008 Sep 22. Oncogene. 2008. PMID: 18806826 Free PMC article.
-
EZH2 methyltransferase and H3K27 methylation in breast cancer.Int J Biol Sci. 2012;8(1):59-65. doi: 10.7150/ijbs.8.59. Epub 2011 Nov 18. Int J Biol Sci. 2012. PMID: 22211105 Free PMC article. Review.
-
Importance of Ezh2 polycomb protein in tumorigenesis process interfering with the pathway of growth suppressive key elements.J Cell Physiol. 2008 Feb;214(2):295-300. doi: 10.1002/jcp.21241. J Cell Physiol. 2008. PMID: 17786943 Review.
Cited by
-
MicroRNAs: Diverse Mechanisms of Action and Their Potential Applications as Cancer Epi-Therapeutics.Biomolecules. 2020 Sep 7;10(9):1285. doi: 10.3390/biom10091285. Biomolecules. 2020. PMID: 32906681 Free PMC article. Review.
-
EZH2-Targeted Therapies in Cancer: Hype or a Reality.Cancer Res. 2020 Dec 15;80(24):5449-5458. doi: 10.1158/0008-5472.CAN-20-2147. Epub 2020 Sep 25. Cancer Res. 2020. PMID: 32978169 Free PMC article. Review.
-
Curcumin analogue CDF inhibits pancreatic tumor growth by switching on suppressor microRNAs and attenuating EZH2 expression.Cancer Res. 2012 Jan 1;72(1):335-45. doi: 10.1158/0008-5472.CAN-11-2182. Epub 2011 Nov 22. Cancer Res. 2012. PMID: 22108826 Free PMC article.
-
Cistrome analysis of YY1 uncovers a regulatory axis of YY1:BRD2/4-PFKP during tumorigenesis of advanced prostate cancer.Nucleic Acids Res. 2021 May 21;49(9):4971-4988. doi: 10.1093/nar/gkab252. Nucleic Acids Res. 2021. PMID: 33849067 Free PMC article.
-
Silencing of microRNA-101 prevents IL-1β-induced extracellular matrix degradation in chondrocytes.Arthritis Res Ther. 2012 Dec 10;14(6):R268. doi: 10.1186/ar4114. Arthritis Res Ther. 2012. PMID: 23227940 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

