ATRX marks the inactive X chromosome (Xi) in somatic cells and during imprinted X chromosome inactivation in trophoblast stem cells
- PMID: 19005673
- PMCID: PMC2877807
- DOI: 10.1007/s00412-008-0189-x
ATRX marks the inactive X chromosome (Xi) in somatic cells and during imprinted X chromosome inactivation in trophoblast stem cells
Abstract
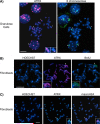
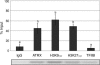
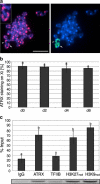
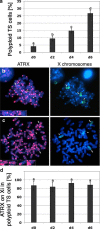
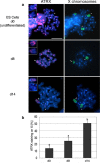
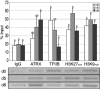
Mammalian X chromosome inactivation (XCI) is an essential mechanism to compensate for dosage imbalances between male and female embryos. Although the molecular pathways are not fully understood, heterochromatinization of the Xi requires the coordinate recruitment of multiple epigenetic marks. Using fluorescence in situ hybridization analysis combined with immunocytochemistry, we demonstrate that the chromatin remodeling protein ATRX decorates the chromatids of a single, late replicating X chromosome in female somatic cells and co-localizes with the bona fide marker of the Xi, macroH2A1.2. Chromatin immunoprecipitation using somatic, embryonic stem (ES) cells and trophoblast stem (TS) cells as model for random and imprinted XCI, respectively, revealed that, in somatic and TS cells, ATRX exhibits a specific association with sequences located within the previously described H3K9me2-hotspot, a region 5' to the X inactive-specific transcript (Xist) locus. While no ATRX-Xi interaction was detectable in undifferentiated ES cells, an enrichment of ATRX was observed after 8 days of differentiation, indicating that ATRX associates with the Xi following the onset of random XCI, consistent with a potential role in maintenance of XCI. These results have important implications regarding a previously described escape from imprinted XCI in ATRX-deficient mice as well as cases of skewed XCI in patients with ATRX syndrome.
Figures

Similar articles
-
Failure of extra-embryonic progenitor maintenance in the absence of dosage compensation.Development. 2012 Jun;139(12):2130-8. doi: 10.1242/dev.076497. Epub 2012 May 9. Development. 2012. PMID: 22573614 Free PMC article.
-
Lineage-specific function of the noncoding Tsix RNA for Xist repression and Xi reactivation in mice.Genes Dev. 2011 Aug 15;25(16):1702-15. doi: 10.1101/gad.16997911. Genes Dev. 2011. PMID: 21852535 Free PMC article.
-
Role of ATRX in chromatin structure and function: implications for chromosome instability and human disease.Reproduction. 2011 Aug;142(2):221-34. doi: 10.1530/REP-10-0380. Epub 2011 Jun 8. Reproduction. 2011. PMID: 21653732 Free PMC article. Review.
-
High-resolution analysis of epigenetic changes associated with X inactivation.Genome Res. 2009 Aug;19(8):1361-73. doi: 10.1101/gr.092643.109. Epub 2009 Jul 6. Genome Res. 2009. PMID: 19581487 Free PMC article.
-
Female-bias in systemic lupus erythematosus: How much is the X chromosome to blame?Biol Sex Differ. 2024 Oct 7;15(1):76. doi: 10.1186/s13293-024-00650-y. Biol Sex Differ. 2024. PMID: 39375734 Free PMC article. Review.
Cited by
-
Lessons from comparative analysis of X-chromosome inactivation in mammals.Chromosome Res. 2009;17(5):659-69. doi: 10.1007/s10577-009-9057-7. Chromosome Res. 2009. PMID: 19802706 Review.
-
XCI in preimplantation mouse and human embryos: first there is remodelling….Hum Genet. 2011 Aug;130(2):203-15. doi: 10.1007/s00439-011-1014-9. Epub 2011 Jun 7. Hum Genet. 2011. PMID: 21647603 Free PMC article. Review.
-
Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between Polycomb complexes PRC1 and PRC2.Nat Commun. 2016 Nov 28;7:13661. doi: 10.1038/ncomms13661. Nat Commun. 2016. PMID: 27892467 Free PMC article.
-
Multiple Roles of dXNP and dADD1-Drosophila Orthologs of ATRX Chromatin Remodeler.Int J Mol Sci. 2023 Nov 18;24(22):16486. doi: 10.3390/ijms242216486. Int J Mol Sci. 2023. PMID: 38003676 Free PMC article. Review.
-
New and Xisting regulatory mechanisms of X chromosome inactivation.Curr Opin Genet Dev. 2012 Apr;22(2):62-71. doi: 10.1016/j.gde.2012.02.007. Epub 2012 Mar 16. Curr Opin Genet Dev. 2012. PMID: 22424802 Free PMC article. Review.
References
-
- Abbondanzo S, Gadi I, Stewart C. Methods Enzymology. vol. 225. Academic Press; San Diego: 1993. Derivation of embryonic stem cell lines; pp. 803–823. - PubMed
-
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. - PubMed
-
- Berube NG, Smeenk CA, Picketts DJ. Cell cycle-dependent phosphorylation of the ATRX protein correlates with changes in nuclear matrix and chromatin association. Hum Mol Genet. 2000;9:539–547. - PubMed
-
- Boggs BA, Cheung P, Heard E, Spector DL, Chinault AC, Allis CD. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat Genet. 2002;30:73–76. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

