Improvement of bacterial transformation efficiency using plasmid artificial modification
- PMID: 19004868
- PMCID: PMC2615632
- DOI: 10.1093/nar/gkn884
Improvement of bacterial transformation efficiency using plasmid artificial modification
Abstract
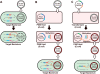
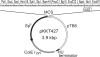
We have developed a method to improve the transformation efficiency in genome-sequenced bacteria, using 'Plasmid Artificial Modification' (PAM), using the host's own restriction system. In this method, a shuttle vector was pre-methylated in Escherichia coli cells, which carry all the putative genes encoding the DNA modification enzymes of the target microorganism, before electroporation was performed. In the case of Bifidobacterium adolescentis ATCC15703 and pKKT427 (3.9 kb E. coli-Bifidobacterium shuttle vector), introducing two Type II DNA methyltransferase genes lead to an enhancement in the transformation efficiency by five orders of magnitude. This concept was also applicable to a Type I restriction system. In the case of Lactococcus lactis IO-1, by using PAM with a putative Type I methyltransferase system, hsdMS1, the transformation efficiency was improved by a factor of seven over that without PAM.
Figures



Similar articles
-
Plasmid artificial modification: a novel method for efficient DNA transfer into bacteria.Methods Mol Biol. 2011;765:309-26. doi: 10.1007/978-1-61779-197-0_18. Methods Mol Biol. 2011. PMID: 21815100
-
Transformation of Bifidobacterium longum with pRM2, a constructed Escherichia coli-B. longum shuttle vector.Plasmid. 1994 Sep;32(2):208-11. doi: 10.1006/plas.1994.1056. Plasmid. 1994. PMID: 7846144
-
Improvement of electroporation-mediated transformation efficiency for a Bifidobacterium strain to a reproducibly high level.J Microbiol Methods. 2019 Apr;159:112-119. doi: 10.1016/j.mimet.2018.11.019. Epub 2018 Dec 5. J Microbiol Methods. 2019. PMID: 30529116
-
Current approaches to genetic modification of marine bacteria and considerations for improved transformation efficiency.Microbiol Res. 2024 Jul;284:127729. doi: 10.1016/j.micres.2024.127729. Epub 2024 Apr 20. Microbiol Res. 2024. PMID: 38663232 Review.
-
Technological advances in bifidobacterial molecular genetics: application to functional genomics and medical treatments.Biosci Microbiota Food Health. 2012;31(2):15-25. doi: 10.12938/bmfh.31.15. Epub 2012 Apr 20. Biosci Microbiota Food Health. 2012. PMID: 24936345 Free PMC article. Review.
Cited by
-
Development of a double-crossover markerless gene deletion system in Bifidobacterium longum: functional analysis of the α-galactosidase gene for raffinose assimilation.Appl Environ Microbiol. 2012 Jul;78(14):4984-94. doi: 10.1128/AEM.00588-12. Epub 2012 May 11. Appl Environ Microbiol. 2012. PMID: 22582061 Free PMC article.
-
Engineering Bacillus subtilis ATCC 6051a for the production of recombinant catalases.J Ind Microbiol Biotechnol. 2021 Jul 1;48(5-6):kuab024. doi: 10.1093/jimb/kuab024. J Ind Microbiol Biotechnol. 2021. PMID: 33734388 Free PMC article.
-
Genetic manipulation of Staphylococci-breaking through the barrier.Front Cell Infect Microbiol. 2012 Apr 12;2:49. doi: 10.3389/fcimb.2012.00049. eCollection 2012. Front Cell Infect Microbiol. 2012. PMID: 22919640 Free PMC article. Review.
-
Controlled gene expression in bifidobacteria by use of a bile-responsive element.Appl Environ Microbiol. 2012 Jan;78(2):581-5. doi: 10.1128/AEM.06611-11. Epub 2011 Nov 11. Appl Environ Microbiol. 2012. PMID: 22081575 Free PMC article.
-
Mechanistic Study of Utilization of Water-Insoluble Saccharomyces cerevisiae Glucans by Bifidobacterium breve Strain JCM1192.Appl Environ Microbiol. 2017 Mar 17;83(7):e03442-16. doi: 10.1128/AEM.03442-16. Print 2017 Apr 1. Appl Environ Microbiol. 2017. PMID: 28115383 Free PMC article.
References
-
- Arber W, Linn S. DNA modification and restriction. Annu. Rev. Biochem. 1969;38:467–500. - PubMed
-
- Tock MR, Dryden DT. The biology of restriction and anti-restriction. Curr. Opin. Microbiol. 2005;8:466–472. - PubMed
-
- Tanaka K, Samura K, Kano Y. Structural and functional analysis of pTB6 from Bifidobacterium longum. Biosci. Biotechnol. Biochem. 2005;69:422–425. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

